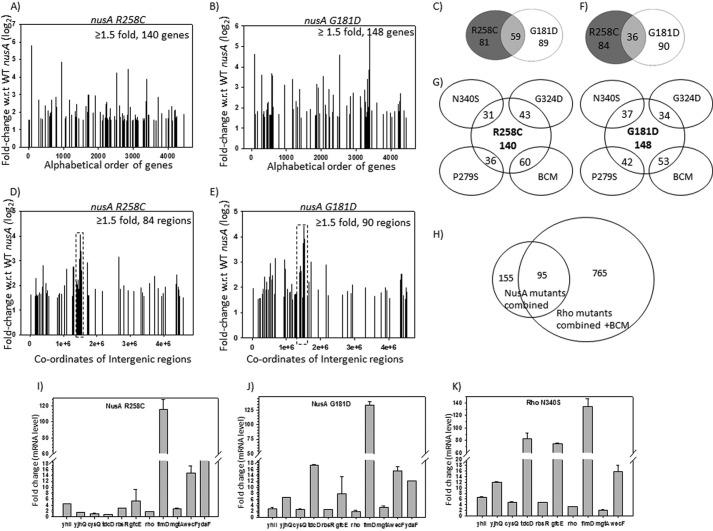
FIGURE 8.
Microarray profiles and their validations by q-PCR. A and B, show the plots of microarray profiles of the coding regions (genes) obtained from MG1655rac+ strains expressing different nusA mutants as indicated. The ratio of the hybridization intensities obtained from WT and different mutants gave the measure of the -fold changes that is expressed in log2 scale as per the convention. Up-regulated genes with ≥1.5-fold changes are shown. Each gene is covered by ∼60 probes, and the averages of the -fold change values obtained from each of the probes were assigned as the -fold change values of each gene. C, Venn diagram showing the number of overlapping genes and operons between the two nusA mutants. D and E, microarray profile plots for the non-coding regions obtained under the same conditions as in A and B. The coordinate at the midpoint of each of the intergenic regions was considered for plotting. F, Venn diagram showing the overlaps between the non-coding regions of the two NusA mutants. G, Venn diagrams showing the numbers of overlapping genes and operons between the NusA (R258C and G181D) and the Rho mutants (N340S, G324D, P279S) and those obtained in the presence of bicyclomycin (BCM). H, overlapping patterns of total number of up-regulated genes obtained in all of the NusA and Rho mutants together with those obtained in the presence of the BCM. I–K, validations of the microarray data of the indicated genes by qRT-PCR. In vivo levels of RNA of the up-regulated genes in the presence of the NusA mutants, G181D (H) and R258C (I), and the Rho mutant, N340S (J). mRNA levels are expressed as -fold changes of the CT values (threshold cycle) obtained from the qRT-PCR as per the convention, details of which are given under “Experimental Procedures.” S.D. values (error bars) were calculated from the two biological replicates. y axis breaks are used to accommodate the wide range of -fold change values for different genes.