Abstract
Dimunito/Dwarf1 (DWF1) is an oxidoreductase enzyme that is responsible for the conversion of C28- and C29-Δ24(28)-olefinic sterols to 24-methyl- and 24-ethylcholesterols. Generally, the reaction proceeds in two steps via the Δ24(25) intermediate. In this study, we characterized the ArDWF1 gene from an expression sequence tag library of Ajuga reptans var. atropurpurea hairy roots. The gene was functionally expressed in the yeast T21 strain. The in vivo and in vitro study of the transformed yeast indicated that ArDWF1 catalyzes the conversion of 24-methylenecholesterol to campesterol. A labeling study followed by GC-MS analysis suggested that the reaction proceeded with retention of the C-25 hydrogen. The 25-H retention was established by the incubation of the enzyme with (23,23,25-2H3,28-13C)-24-methylenecholesterol, followed by 13C NMR analysis of the resulting campesterol. Thus, it has been concluded that ArDWF1 directly reduces 24-methylenecholesterol to produce campesterol without passing through a Δ24(25) intermediate. This is the first characterization of such a unique DWF1 enzyme. For comparison purposes, Oryza sativa DWF1 (OsDWF1) was similarly expressed in yeast. An in vivo assay of OsDWF1 supported the generally accepted two-step mechanism because the C-25 hydrogen of 24-methylenecholesterol was eliminated during its conversion to 24-methylcholesterol. As expected, the 24-methylcholesterol produced by OsDWF1 was a mixture of campesterol and dihydrobrassicasterol. Furthermore, the 24-methylcholesterol contained in the Ajuga hairy roots was determined to be solely campesterol through its analysis using chiral GC-MS. Therefore, ArDWF1 has another unique property in that only campesterol is formed by the direct reduction catalyzed by the enzyme.
Keywords: biosynthesis, enzyme mechanism, plant biochemistry, reductase, sterol
Introduction
In vertebrates, cholesterol is the major sterol, whereas, in higher plants, commonly occurring sterols are C29 sterols, sitosterol (1) and stigmasterol, and a C28 sterol, campesterol (2). Campesterol is frequently accompanied by dihydrobrassicasterol (3) (1, 2). Latter steps in phytosterol biosynthesis have received considerable attention because of their intriguing chemistry, biochemistry, and biology. Recent studies through genetic mutations have uncovered an essential role of these phytosterols at the cellular level in hormone signaling, organized divisions, and embryo patterning (3).
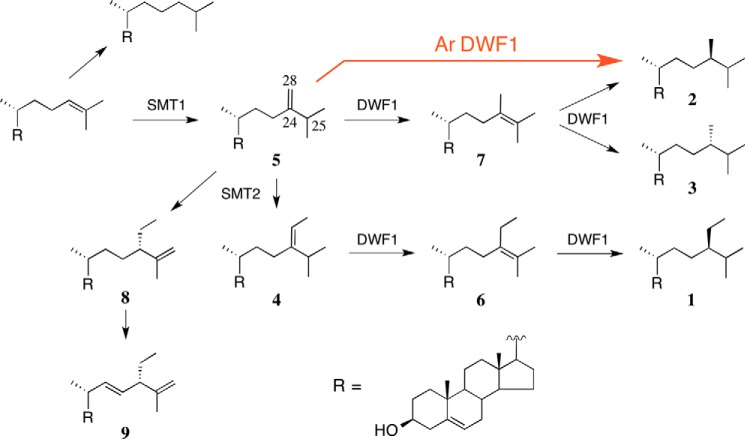
It is generally regarded that these typical plant sterols are biosynthesized from precursor Δ24(28)-olefinic sterols, i.e. isofucosterol (4) and 24-methylenecholesterol (5), in two steps: a double-bond isomerization from Δ24(28) to Δ24(25) (leading to 24-ethyldesmosterol (6) and 24-methyldesmosterol (7)) and a reduction of the Δ24(25) intermediates to the saturated C29 and C28 sterols (1 and 2/3). Interestingly, the reduction of 24-ethyldesmosterol affords only sitosterol, whereas the reduction of 24-methyldesmosterol yields a C-24 epimeric mixture of campesterol and dihydrobrassicasterol (1, 2). The two-step mechanism has been proposed mainly on the basis of the metabolic behavior of the C-25 hydrogen of the Δ24(28)-olefinic sterol, which is eliminated during plant sterol biosynthesis (4–6). The C28-Δ24(28)-olefinic sterol (e.g. 24-methylenecholesterol) is produced by the action of sterol methyltransferase (SMT-1) to a Δ24(25) intermediate (cycloartenol is reportedly the most preferable substrate (7) and then followed by hydrogen migration from C-24 to C-25 and elimination of the C-28 hydrogen (8–10). The homologous C29-Δ24(28)-olefinic sterol (e.g. isofucosterol) is produced by the action of SMT-2 to a C28-Δ24(28)-olefinic intermediate (24-methylenelophenol is reportedly the most preferred substrate), followed by elimination of the C-28 hydrogen (Fig. 1).
FIGURE 1.
Biosynthetic pathway of 24-methyl and 24-ethyl sterols in higher plants. This study determined that ArDWF1 catalyzes the direct reduction of a Δ24(28) sterol (red arrow) as opposed to the typical two-step mechanism that includes double-bond migration. To simplify the discussion, all structures are depicted as having 3β-hydroxy-Δ5 nuclei. However, this does not necessarily mean that the reactions proceed as the nuclei structure.
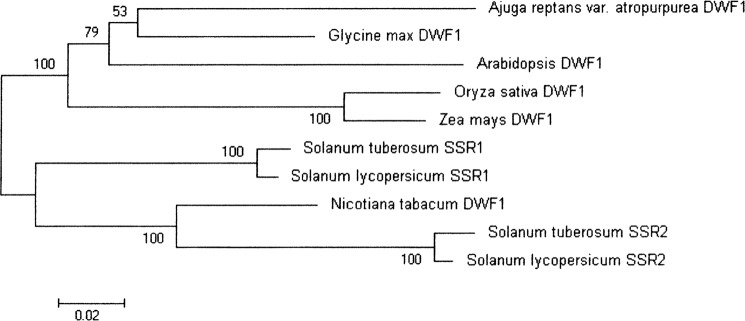
The Dimunito/Dwarf1 (DWF1) gene, which encodes an oxidoreductase with an FAD binding domain, was first cloned from Arabidopsis thaliana (AtDWF1) (11), and it was subsequently indicated that this gene is involved in both the double-bond isomerization and the reduction step (12). A. thaliana and Oryza sativa (13) each have a single DWF1 gene, whereas tomato and potato have two DWF1 ortholog genes (SSR1 and SSR2) (14). Therefore, it is highly likely that DWF1 of A. thaliana as well as O. sativa catalyzes the formation of sitosterol as well as campesterol and dihydrobrassicasterol. It has been recently reported that tomato SSR1 has a substrate specificity for 24-methylenecholesterol, affording 24-methylcholesterol, whereas SSR2 preferentially reduces a Δ24(25)-sterol (cycloartenol/desmosterol) to produce cycloartenol/cholesterol (14).
It has been reported that the double-bond migration of the C29-Δ24(28)-olefinic sterol proceeds through a syn-SE2′ mechanism (15). Furthermore, the reduction of the C28-Δ24(25)-olefinic sterol proceeds in a trans-addition manner wherein the hydrogen atoms are introduced from both the 24-Si, 25-Re, and the 24-Re, 25-Si faces to give campesterol and dihydrobrassicasterol, respectively, in O. sativa cell culture and in the Catharanthus roseus callus (16, 17). It has also been indicated that 24-ethyldesmostrol is reduced to give sitosterol in a trans-addition manner, wherein the hydrogen atoms are introduced only from the 24-Si, 25-Re face (16, 18).
In the study of phytoecdysteroid biosynthesis, we used the hairy roots of Ajuga reptans var. atropurpurea (Labiatae family) (19–22). Ajuga plants have unique sterol profiles in which clerosterol (8) and dehydroclerosterol (9) are the major sterols (20, 23). These sterols are formed by the trans-methylation of a C28-Δ24(28) intermediate followed by a 1,2-hydrogen shift from C-25 to C-24 and deprotonation at C-27 (24). Ajuga plants also contain C29-, C28-, and C27-phytoecdysteroids, including cyasterone and 20-hydroxyecdysone. We reported that, in Ajuga hairy roots, cholesterol is a precursor of 20-hydroxyecdysone (19), whereas cyasterone is biosynthesized from clerosterol (25). To accelerate our study on the biosyntheses of sterols and ecdysteroids in Ajuga plants, we recently generated an expression sequence tag library of Ajuga hairy roots (26) from which a DWF1 ortholog gene (designated ArDWF1) was identified.
In this paper, we report a unique reaction mechanism catalyzed by ArDWF1, which is distinct from the generally accepted two-step mechanism. O. sativa Dwarf1 (OsDWF1) was also studied as the basis for comparison.
Experimental Procedures
Analytical Method
The 1H and 13C NMR spectra were recorded using a Bruker DRX-500 (500 MHz for 1H and 125 MHz for 13C) spectrometer in CDCl3 (99.8% atom enriched, Isotec) as the solvent. The 1H chemical shifts are reported in reference to the signal of tetramethylsilane (δH 0.00), whereas 13C chemical shifts are in reference to the solvent signals (δC 77.0 for CDCl3).
GC-MS Analysis
A mass spectrometer (JMS-AM SUN200, JEOL) was connected to a gas chromatograph (6890A, Agilent Technologies) with an electron ionization of 70 eV, a source temperature of 250 °C, a DB-1ms column of 30 m × 0.25 mm, a film thickness of 0.25 μm (J&W Scientific), and an injection temperature of 250 °C. The following column temperature program was used: 80 °C for 1 min, an increase to 300 °C at a rate of 20 °C/min, and held at 300 °C for 12 min. The interface temperature was 280 °C, the carrier gas was helium, and the flow rate was 1 ml/min. Splitless injections were performed.
GC-MS analysis was performed using a chiral column, HP-Chiral 20β (30 m × 0.25 mm, film thickness of 0.25 μm, Agilent) with the following column temperature program: held at 80 °C for 1 min, raised to 250 °C at a rate of 20 °C/min, and held at 250 °C for 71 min. Authentic campesterol and dihydrobrassicasterol were injected as references.
HPLC Analysis
An LC-6A apparatus (Shimadzu) equipped with an SPD-6A UV detector (215 nm) and a Shim-Pack CLC-ODS column (15 cm × 6 mm inner diameter, Shimadzu) was used.
cDNA Cloning of ArDWF1
The details of the preparation of an expression sequence tag library from the Ajuga hairy roots will be reported elsewhere (26). Keyword searches of the annotated expression sequence tags identified only one DWF1 contig.2 The sequence data allowed the design of primers with the following sequences: 5′-AGTCGGATCCATGTCAGATCTTGAGGTCCCCC-3′ and 5′-AGTCGCTAGCTTAGGC AACTTCAGCATCGGGAG-3′. The open reading frame of ArDWF1was amplified by PCR (initial denaturing for 2 min at 98 °C; 35 cycles of 94 °C for 10 s, 55 °C for 5 s, and 72 °C for 30 s; and a final extension reaction for 5 min at 72 °C) using PrimeSTAR max with the template (a cDNA library that was constructed from Ajuga hairy roots) and the primers (described above). The PCR product was ligated into the galactose-inducible yeast expression vector pESC-LEU using the BamHI and NheI sites included in the primers. The identity of the inserted cDNA was confirmed by sequencing.
Yeast Transformation by ArDWF1 and OsDWF1
The yeast T21 strain that produces 24-methylenecholesterol, which was constructed by Sawai et al. (14), was transformed with the plasmids or the empty pESC-LEU vector as a control using the Frozen-EZ Transformation II kit (Zymo Research, CA). The transformants were selected using a synthetic complete medium (Takara Bio) containing 2% (w/v) glucose supplemented with -His/–Leu or -Ura/–His DO supplement (Takara Bio) for 2 days at 30 °C.
cDNA Cloning of OsDWF1
Seeds of the O. sativa L. cv. Nipponbare (Nihon Nouken, Inc.) were sterilized with running water at 25 °C for 12 h, and then vernalization was done at 4 °C under dark conditions for 12 h. The seeds were germinated on a nylon net floating on reverse osmosis-purified water. The seedlings were grown at 28 °C using a photoperiodic cycle of 12 h light/12 h dark for 14 days. The total RNA of the seedlings was extracted using the RNeasy plant mini kit (Qiagen) according to the specifications of the manufacturer. The extract was treated with DNase using the RNase-free DNase set (Qiagen) and purified using the RNeasy kit. First-strand cDNA synthesis was performed using the SuperScript III first-strand synthesis system for RT-PCR kit (Invitrogen) utilizing 1 μg of total RNA. A PCR fragment of OsDWF1 was amplified by PCR with the primer set 5′-ATGGCAGATCTGCAGGAGCC-3′ and 5′-TTACGCCTCATCAGCGTAGGC-3′ and template rice cDNA. The insert was cloned into the Gateway entry vector pENTR/D-TOPO (Life Technologies) and then into the yeast expression vector pYES-DEST52 (Life Technologies) with the Gateway LR clonase (Life Technologies) reaction. The obtained plasmid and empty vector were introduced into the yeast T21 strain.
In Vivo Assay with ArDWF1- and OsDWF1-expressing Yeasts
The transformed yeast cells were inoculated into 5 ml of the same medium and cultured for 2 days at 30 °C. The cells were harvested by centrifugation and resuspended in 5 ml of new medium containing 2% (w/v) galactose instead of glucose. After incubation for another 2 days at 30 °C, the cells were disrupted by vortexing with glass beads (0.5-mm diameter) and extracted with ethyl acetate (EtOAc) (5 ml × 3). The organic phase of the extraction was dried over Na2SO4 and concentrated in vacuo. A part of the residue was trimethylsilylated with N-methyl-N-trimethylsilyltrifluoroacetamide (Sigma-Aldrich) at 80 °C for 30 min and analyzed by GC-MS.
Determination of the C-24 Stereochemistry of 24-Methylcholesterol Produced in Vivo by 1H NMR Spectroscopy
The cultivation of ArDWF1-expressing yeast was carried out in 500 ml of synthetic complete medium containing 2% (w/v) glucose supplemented with the -His/–Leu DO supplement. The production cultures were inoculated with 5% (w/v) of the preculture with the same medium. After incubation of the culture for 2 days at 30 °C and 150 rpm, the cells were harvested and resuspended in the same volume of fresh medium containing 2% (w/v) galactose instead of glucose. Further incubation occurred for 2 days at 30 °C. Then the harvested cells were lyophilized, ground with a mortar and pestle, and extracted with EtOAc (10 ml × 3). The concentrated residue was chromatographed over silica gel with hexane-EtOAc (4:1) to separate the sterol fraction. The fraction was further separated by reverse-phase HPLC (solvent, MeOH; flow rate, 1.0 ml/min; retention time, 16.4 min) to yield 24-methylcholesterol, which was analyzed by 1H NMR.
A large-scale incubation (1.5 liters of the medium) of yeast expressing OsDWF1 was carried out in the same manner as for ArDWF1-expressing yeast. The harvested cells were treated as described above to give 24-methycholesterol, which was analyzed by 1H NMR.
Determination of the C-24 Stereochemistry of 24-Methylcholesterol Contained in Ajuga Hairy Roots and the O. sativa Callus by Chiral GC-MS
The hairy roots of A. reptans var. atropurpurea were maintained as described previously (18). The hairy roots were transferred to a 100-ml Erlenmeyer flask containing 1 × Murashige and Skoog medium (30 ml) supplemented with 3% (w/v) sucrose and incubated at 25 °C on a rotary shaker (70 rpm) in the dark for 3 weeks. The hairy roots were harvested, washed with water, and lyophilized (dry weight, 0.5 g). A portion of the dried hairy roots (50 mg) was ground and extracted with CHCl3-methanol (1:1, 2 ml). After the addition of Celite to the extract, the solvents were removed under reduced pressure. The adsorbed sample was placed in a Sep-Pak Vac (silica, 500 mg/6 ml, Waters) and eluted with hexane-EtOAc (2:1, 8 ml) and then CHCl3-methanol (1:1, 8 ml). The hexane-EtOAc eluent was concentrated, and the residue was saponified with 4 m KOH (1 ml) in ethanol for 1 h at 80 °C and then diluted with water (2 ml). The CHCl3-methanol eluent was concentrated, and the residue was hydrolyzed with 1 ml each of methanol and 4 m HCl for 1 h at 80 °C. Both reaction mixtures were extracted with hexane (2 ml × 2), and the hexane layers were combined and concentrated to dryness. A portion of the residue was trimethylsilylated and analyzed by GC-MS using a chiral column.
O. sativa cell culture was maintained as described previously (16). Cultured cells of O. sativa were transferred to a 100-ml Erlenmeyer flask containing N6 liquid medium (30 ml) supplemented with 2% (w/v) sucrose, N6 vitamin (0.3 ml), proline (2.8 g/liter), casein hydrolysate (300 mg/liter), 2,4-dichlorophenoxyacetic acid (2 mg/liter), and myo-inositol (100 mg/liter) and grown on a rotary shaker (70 rpm) at 25 °C in the dark for 14 days. The cells harvested by filtration were processed as described for the Ajuga hairy roots, and the hexane extract was analyzed similarly.
Administration of (23,23,25-2H3,28-13C)-24-methyleneholesterol to Ajuga Hairy Roots and the O. sativa Callus
Ajuga hairy roots were cultured in a 100-ml Erlenmeyer flask for 2 weeks as described above. A sterilized solution of (23,23,25-2H3,28-13C)-24-methyleneholesterol (2.0 mg) and methyl-β-cyclodextrin (100 mg) in H2O (2.0 ml) was added to the liquid medium, and incubation was continued for another 2 weeks. The hairy roots were harvested and treated as described above, and the hexane extract was trimethylsilylated and analyzed by GC-MS.
Similarly, the labeled 24-methyleneholesterol (2.0 mg) was added to O. sativa cells that were grown in a 100-ml Erlenmeyer flask for 2 weeks. The cell culture was incubated for another 2 weeks, and the hexane extract was obtained as described above. The residue was analyzed by GC-MS after trimethylsilylation.
Incubation of Labeled Sterol Substrates with ArDWF1-expressing Yeast Homogenate
The transformant was cultured in synthetic complete medium (250 ml) containing 2% (w/v) glucose supplemented with the -His/–Leu DO supplement for 2 days at 30 °C. The cells were harvested and resuspended in new medium (250 ml) containing 2% (w/v) galactose instead of glucose. After incubation for one more day at 30 °C, the cells were harvested and resuspended in 100 mm Tris-HCl buffer (5 ml, pH 7.23) containing 0.1 mm dithiothreitol and 1 mm EDTA. The cells were lysed using a bead beater (Wakenyaku) with glass beads (0.5-mm diameter, 5 ml for the cell amount of ∼10 ml) for 30 s × 4 with 4-min cooling intervals. The resulting cell homogenate was centrifuged at 1000 × g for 5 min at 4 °C. The supernatant was used for an enzymatic assay (5 ml/substrate). The assay mixture contained 30 mm nicotinamide, 3.5 mm NADPH, 3.5 mm FAD, 0.5 mg/ml bovine serum albumin, and 168 μm (23,23,25-2H3,28-13C)-24-methylenecholesterol and (28,28,28-2H3,28-13C)-24-methyldesmosterol. These two compounds were prepared as 420 μm stock suspensions in Tris-HCl (pH 7.23) containing 1.25% (w/v) methyl-β-cyclodextrin (Wako Pure Chemical Industries). The assay solution was incubated for 3 h at 30 °C. The reaction was terminated by adding EtOAc (5 ml) and vortexing vigorously. The whole mixture was extracted with EtOAc (5 ml × 3). The organic layer was concentrated to dryness, and the residue was analyzed by GC-MS as the TMS ether.
A large-scale homogenate (60 ml) of ArDWF1-expressing yeast was prepared and incubated with (23,23,25-2H3,28-13C)-24-methylenecholesterol (4.0 mg) as described above. The EtOAc extract was chromatographed over silica gel eluting with hexane-EtOAc (4:1) to give a sterol-enriched fraction. This fraction was separated by HPLC to isolate 24-methylcholesterol, which was analyzed by 2H-decoupled 13C NMR spectroscopy.
Incubation of Labeled Sterol Substrates with OsDWF1-expressing Yeast Homogenate
The yeast homogenate was prepared in the same manner as described for the homogenate of the ArDWF1-expressing yeast. (23,23,25-2H3,28-13C)-24-methylenecholesterol and (28,28,28-2H3,28-13C)-24-methylenecholesterol were also incubated with the homogenate for 24 h, as described for the ArDWF1-expressing yeast homogenate. The EtOAc extract was passed through a Sep-Pak Vac (C18, 500 mg/6 mg, Waters) using MeOH. The eluate was concentrated and purified by HPLC (solvent, MeOH; flow rate, 1.0 ml) to obtain 24-methylcholesterol, if any was present. This was analyzed by GC-MS as the TMS ether.
Transfection of S2 Cells
The cDNA of OsDWF1 was cloned into a pUAST vector using its EcoRI and NotI sites. An empty vector of pUAST was used as a negative control. Drosophila melanogaster S2 cells were cultured in Schneider's Drosophila medium (Life Technologies) with 10% (v/v) heat-inactivated fetal calf serum and a penicillin-streptomycin solution (Wako). The S2 cells were transfected with the vector containing the OsDWF1 and Actin5c-GAL4 constructs (a gift from R. Niwa) in 60-mm dishes using Effectene transfection reagent (Qiagen) as described previously (27).
Incubation of (23,23,25-2H3,28-13C)-24-Methylenecholesterol with Transfected S2 Cells
Forty-eight hours after the transfection of S2 cells, the medium was replaced with fresh medium (2 ml) containing the labeled 24-methylenecholesterol (2.0 mg) and methyl-β-cyclodextrin (0.5% w/v). After incubation at 25 °C for 24 h, the medium together with the cells was collected and mixed with EtOAc (2 ml). The mixture was vortexed and centrifuged. The supernatant was separated. The residue was extracted with EtOAc once more, and the combined EtOAc layer was concentrated. The residue was analyzed by GC-MS as the TMS ether.
Steroidal Compounds
24-Methylenecholesterol was prepared as reported previously (28). (23,23,25-2H3,28-13C)-24-methylenecholesterol was prepared using a slight modification of the synthesis of (23,23,25-2H3)-24-methylenecholesterol (28) in which 13C-methyltriphenylphosphonium iodide (Tokyo Chemical Industry) was used in place of its non-labeled counterpart. (28,28,28-2H3,28-13C)-24-methyldesmosterol was prepared from 24-oxocholesterol 3-O-t-butyldimethylsilyl ether (29) in three steps: a Grignard addition of 13CD3-methylmagnesium iodide prepared from 13CD3I (Cambridge Isotope Laboratories), a dehydration of the resulting tertiary alcohol with POCl3 in pyridine, and a desilylation step with tetrabutylammonium fluoride/tetrahydrofuran (1.0 m solution). HPLC was used for the final separation (solvent, MeOH; flow rate, 1.2 ml/min; tR, 15.6 min) of the Δ24(25)-olefinic isomer from Δ23(24)- and Δ24(28)-olefinic isomers. The compound was obtained as white needles, melting point 141–142 °C (crystallized from MeOH), 1H-NMR (500 MHz, CDCl3) δ: 0.68 (s, 18-H3), 0.96 (d, J = 7.6 Hz, 21-H3), 1.01 (s, 19-H3), 1.625 (brs, 26-H3), 1.635 (brs, 27-H3), 3.51 (m, 3-H), 5.35 (m, 6-H), 13C-NMR (125 MHz, CDCl3) intense signal at δ 11.9 (C-18), 17.6 (heptet, J = 18.8 Hz, C-28), 18.8 (C-21), 19.4 (C-19), 20.0 (d, J = 3.8 Hz, C-26), 20.5 (d, J = 5.0 Hz, C-27), 21.1 (C-11), 23.3 (C-15), 28.2 (C-16), 31.0 (d, J = 3.8 Hz, C-23), 31.7 (C-2), 31.9 (C-7, C-8), 34.3(C-22), 36.0 (C-20), 36.5 (C-5), 37.3 (C-1), 39.8 (C-1), 42.3 (C-4, C-13), 121.7 (C-6), 123.3 (C-25), 128.3 (d, J = 43.8 Hz, C-25), 140.8 (C-8) (30). Campesterol and dihydrobrassicasterol were prepared as reported previously (31).
Results
Identification of the ArDWF1 Gene
The protein sequence of ArDWF1 and comparison with those of known DWF1 genes are shown in Figs. 2 and 3. ArDWF1 and OsDWF1 were found to have an FAD binding domain and a transmembrane region; these are also found in AtDWF1 (12).
FIGURE 2.
Alignment of the amino acid sequences of ArDWF1, OsDWF-1, and AtDWF1. The red bar represents the transmembrane region, and the blue bar indicates the FAD binding domain (deduced by SOSUI and PROSITE, respectively). The Ca2+/calmodulin binding region is shown in the blue square. The red square indicates the amino acid that is peculiar to ArDWF1. ArDWF1, A. reptans var. atropurpurea DWF1; GmDWF1, G. max DWF1; AtDWF1, A. thaliana DWF1; OsDWF1, O. sativa DWF1; ZmDWF1, Z. mays DWF1.
FIGURE 3.
Phylogenetic tree showing the relationship of various plant DWF1s. The phylogenetic tree was generated on the basis of the entire amino acid sequences of the proteins using the ClustalW program.
ArDWF1- and OsDWF1-expressing Yeast T21 Cells Produce 24-Methylcholesterol in Vivo
It is expected that DWF1 catalyzes the conversion of the endogenous 24-methylenecholesterol to 24-methylcholesterol when the yeast T21 strain is further transformed with the DWF1 gene. We used this particular yeast transformant in this study because exogenously added 24-methylenecholesterol would have difficulty entering the yeast cells. As expected, GC-MS analysis clearly indicated that ArDWF1-expressing yeast produced 24-methylcholesterol in vivo, whereas the vector control showed a sole peak of 24-methylenecholesterol (Fig. 4). OsDWF1-expressing yeast produced 24-methylcholesterol as well but with a 3-fold less efficiency (estimated by comparing peak integration) than ArDWF1-expressing yeast.
FIGURE 4.
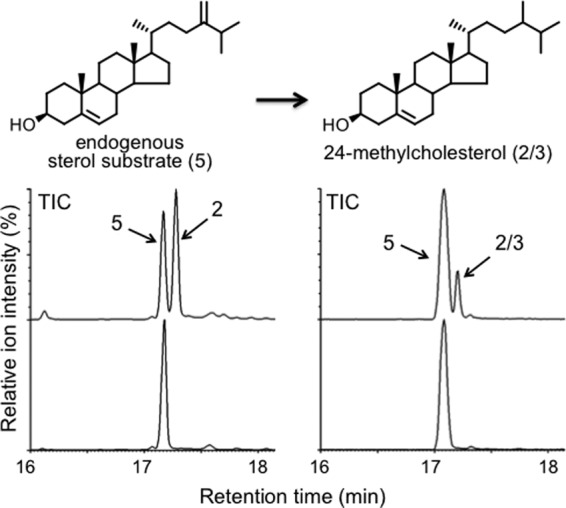
In vivo expression of ArDWF1 and OsDWF1 in yeast. GC-MS analysis (total ion chromatogram (TIC)) of sterols produced by ArDWF1 (left panel) and OsDWF1 (right panel) expressing yeast T21 cells. The peak at tR = 17.24 min was identified as 24-methylcholesterol by direct comparison with campesterol in terms of retention time and mass spectrum. 24-Methylcholesterol produced by ArDWF1 was subsequently found to be campesterol, whereas that obtained by OsDWF1 was determined to be a mixture of campesterol and dihydrobrassicasterol (see text).
The C-24 Configuration of 24-Methylcholesterol
It is known that campesterol and dihydrobrassicasterol can be distinguished by high-resolution 1H-NMR spectroscopy. The 1H-NMR analyses of the HPLC-purified 24-methylcholesterols, which were obtained in a large-scale incubation, unambiguously indicated that ArDWF1 produced campesterol (2) as a single product, whereas OsDWF1 produced an approximately 2:1 mixture of campesterol (2) and dihydrobrassicasterol (3) (Fig. 5). Because the formation of campesterol as a single stereoisomer by ArDWF1 was rather unexpected, the C-24 stereochemistry of the 24-methylcholesterol of Ajuga hairy roots and the O. sativa callus was further investigated.
FIGURE 5.
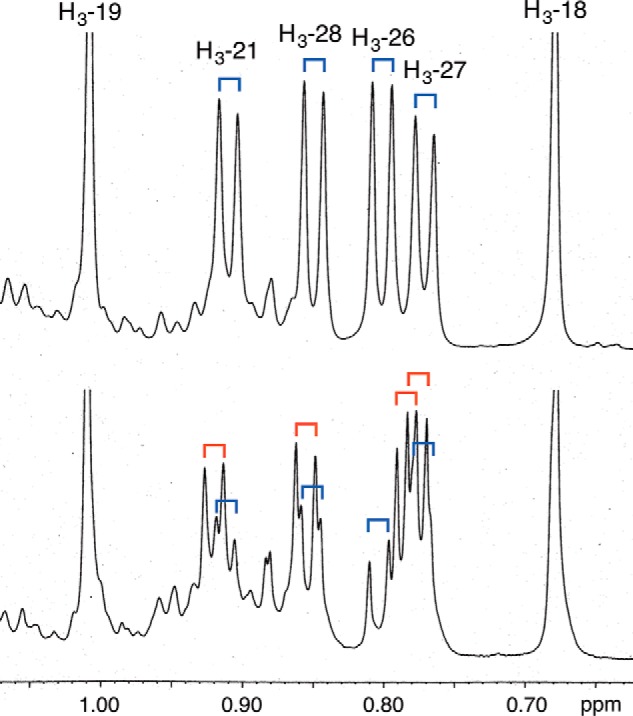
Shown is the determination of the C-24 configuration of 24-methylcholesterol obtained from yeast transformed with ArDWF1 (top) and OsDWF1 (bottom) by 1H NMR. Upfield regions of the spectra are shown. The doublet signals indicated by blue and red correspond to those of campesterol and dihydrobrassicasterol, respectively.
Our preliminary GC-MS study indicated that 24-methylcholesterol consisted of only approximately 0.5–1% of the sterol fraction of Ajuga hairy roots. Thus, we sought a method that could be applied in this case and found that GC-MS analysis using a chiral capillary column was applicable. The analysis indicated that Ajuga hairy roots contained only campesterol as its 24-methylcholesterol, whereas the O. sativa callus contained an ∼1:1 mixture of campesterol and dihydrobrassicasterol, as shown in Fig. 6. The results qualitatively agreed with those of the in vivo experiments. Therefore, it is conceivable that ArDWF1 functions as the sole enzyme responsible for the biosynthesis of 24-methylcholesterol in the Ajuga plant.
FIGURE 6.
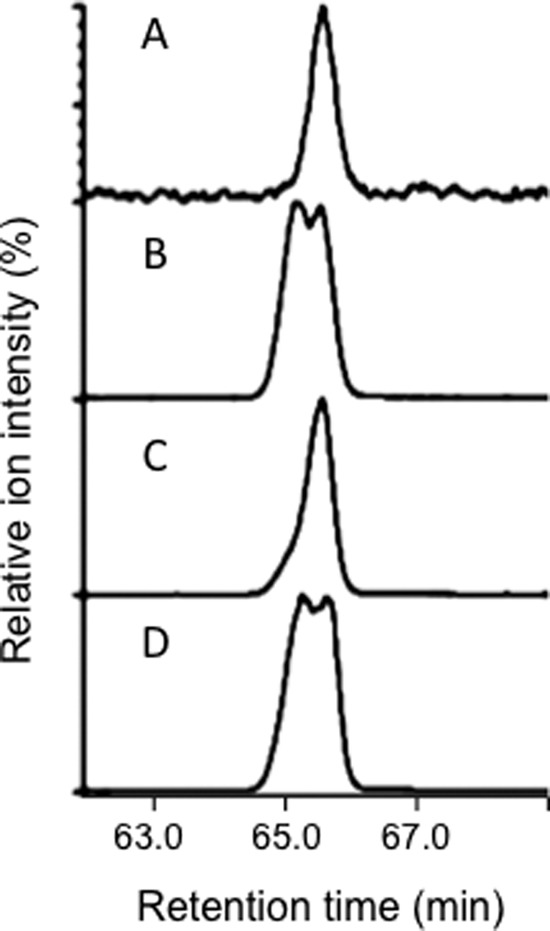
The C-24 configuration of 24-methylcholesterol as determined by GC-MS (total ion chromatogram) using a chiral capillary column. A, from Ajuga hairy roots. B, from the Oryza sativa callus. C, authentic campesterol. D, a 1:1 mixture of authentic dihydrobrassicasterol and campesterol.
Conversion of (23,23,25-2H3,28-13C)-24-Methylenecholesterol to 24-Methylcholesterol in Ajuga Hairy Roots and the O. sativa Callus
The feeding experiments of the labeled substrate were carried out to provide insights into the reaction mechanism of the reductive conversion, focusing on the metabolic fate of H-25. GC-MS analysis (Fig. 7) of the biosynthesized 24-methylsterol (campesterol) fed to Ajuga hairy roots displayed its molecular ion at m/z 476 [M]+. Non-labeled campesterol, which was originally present in the homogenate, showed the molecular ion at m/z 472 [M]+. This result indicated that, during the reductive conversion, the hydrogen atom at C-25 was retained, most likely at the original C-25 position. In contrast, the 24-methylcholesterol obtained from O. sativa callus showed its molecular ion at m/z 475 [M]+, which indicated a loss of one of the deuterium atoms, presumably 25-2H, during the conversion. The finding agreed with the generally accepted two-step mechanism involving the isomerization of Δ24(28) to Δ24(25), which requires elimination of H-25.
FIGURE 7.
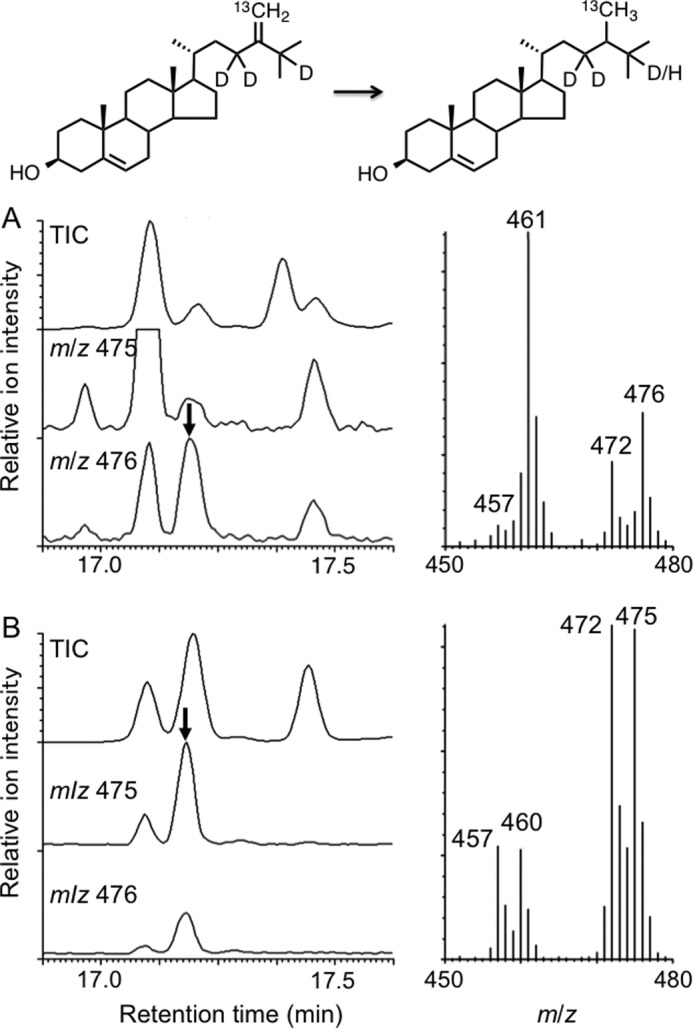
A and B, GC-MS analysis of the 24-methylcholesterol obtained upon feeding (23,23,25-2H3,28-13C)-24-methylenecholesterol to Ajuga hairy roots (A) and Oryza sativa callus (B). The mass spectra were recorded at the peak top of the labeled 24-methylcholesterol indicated by the arrow. The molecular ion peak at m/z 476 for the ArDWF1 product indicated that all three deuterium atoms were retained during the conversion, whereas the molecular ion peak at m/z 475 for the OsDWF1 product implies that one of the deuterium atoms, most likely 25-2H, was lost during the transformation. TIC, total ion chromatogram.
In Vitro Assay of ArDWF1 Using Transformed Yeast Homogenate
The incubation of (23,23,25-2H3,28-13C)-24-methylenecholesterol with a cell-free homogenate (1500 × g supernatant supplemented with NADPH and FAD) of ArDWF1-expressing yeast followed by GC-MS analysis of the product indicated that the substrate was converted into 24-methylcholesterol with good efficiency (Fig. 8). Furthermore, the MS spectrum of the product clearly showed that three deuterium atoms were retained in the molecule. The retention of 25-2H was inconsistent with the two-step mechanism. (28,28,28-2H3,28-13C)-24-Methyldesmosterol was not converted to 24-methylcholesterol using the same homogenate (data not shown).
FIGURE 8.
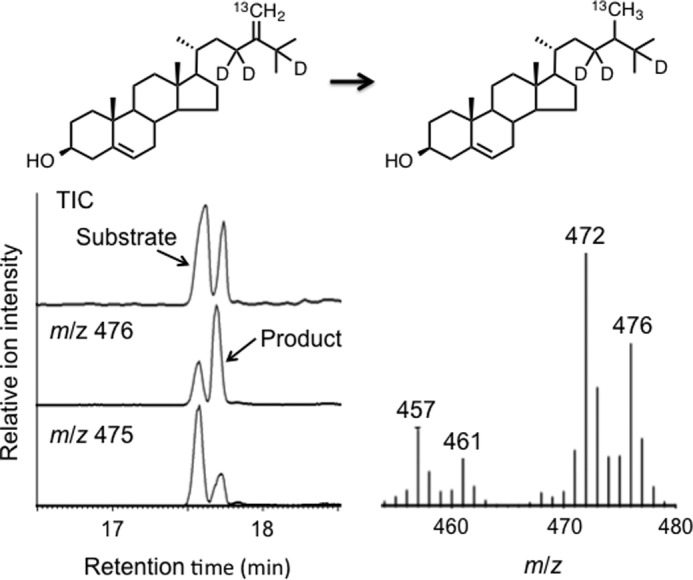
GC-MS analysis (mass chromatograms) of the reaction product obtained upon incubation of (23,23,25-2H3)-24-methylenecholesterol with ArDWF1-expressing yeast homogenate. The mass spectrum was recorded at the peak top (tR = 17.70) of the labeled product. The molecular ion peak at m/z 476 for the ArDWF1 product indicated that all the three deuterium atoms were retained during the conversion. TIC, total ion chromatogram.
Mechanism of the Conversion of 24-Methylenecholesterol to 24-Methylcholesterol with ArDWF1
It was found that H-25 of 24-methylenecholesterol was retained during its conversion to 24-methylcholesterol with ArDWF1, whereas it was eliminated when the reaction was done by OsDWF1. On the basis of the above results, we proposed a direct reduction mechanism without involving the double-bond isomerization for ArDWF1. However, one may question the possibility of the double-bond isomerization concomitant with 1,3-hydrogen migration from C-25 to C-28. In this case, the observed retention of H-25 cannot necessarily eliminate the Δ24(25) intermediate because the hydrogen atom originally located at C-25 can reside at C-28 after the reaction. The 2H-decoupled 13C NMR spectrum of 24-methylcholesterol (campesterol) derived from (23,23,25-2H3,28-13C)-24-methylenecholesterol exhibited an enriched C-28 signal at δ 15.30 ppm (Fig. 9). The data unambiguously indicated that C-28 was not substituted with any deuterium atom. If the deuterium atom migrated from C-25 to C-28, then the C-28 signal should resonate at around δ 15.00 because of a one-bond deuterium isotope shift. The very small upfield shift of C-28 (0.06 ppm from non-labeled C-28) could be attributed to the three-bond deuterium isotope shift of 23,23,25-2H3. We concluded that the conversion of 24-methylenecholesterol to campesterol by ArDWF1 did not pass through a Δ24(25) intermediate.
FIGURE 9.
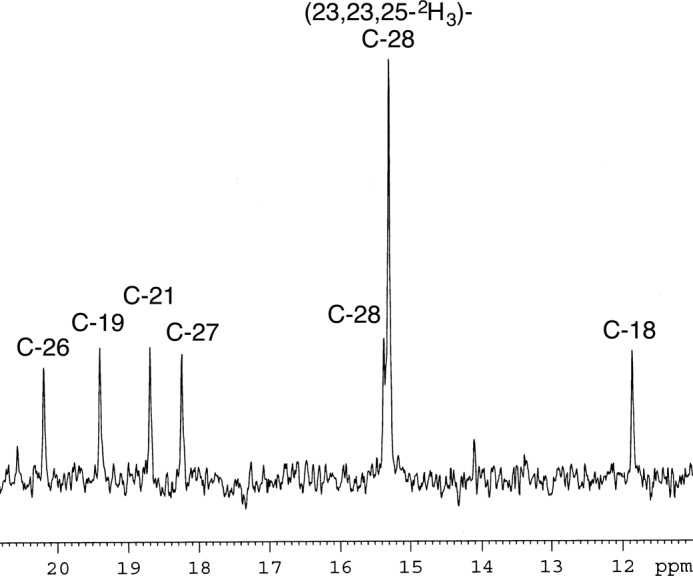
13C NMR analysis of the campesterol obtained upon incubation of (23,23,25-2H3,28-13C)-24-methylenecholesterol with ArDWF1-expressing yeast homogenate. The C-28 signal that originated from the substrate is exhibited at δ 15.30 as an intense peak.
In Vitro Assay of OsDWF1 Using Transformed Yeast Homogenate
(23,23,25-2H3,28-13C)-24-methylenecholesterol was similarly incubated with a cell-free homogenate prepared from OsDWF1-transformed yeast. GC-MS analysis of the incubation product indicated no formation of 24-methylcholesterol (data not shown). The result was markedly different from that with ArDWF1-transformed yeast. In contrast, (28,28,28-2H3,28-13C)-24-methyldesmosterol was converted to 24-methylcholesterol in a few percent yield. The mass chromatogram tracing the m/z 426 ion (Fig. 10) clearly showed this enzymatic conversion. The GC-MS data indicated that the three deuterium atoms at C-28 were retained during the reduction. It is conceivable that the double-bond isomerization step is disturbed for some reason in the cell-free homogenate, which led to the failure of the in vitro conversion of 24-methylenecholesterol to 24-methylcholesterol. In this context, it has been reported that AtDWF1 is a Ca2+/calmodulin-binding protein and that the binding is critical for the catalysis of DWF1 (32).
FIGURE 10.
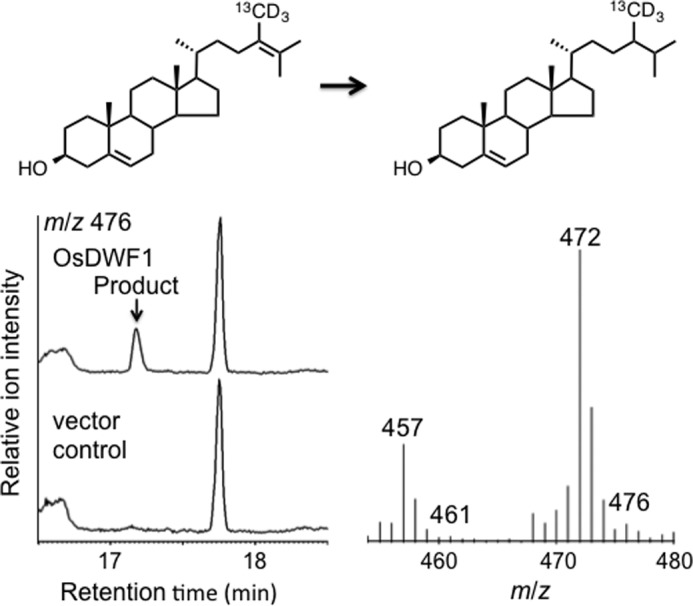
Shown is the GC-MS analysis (mass chromatograms of m/z 476 ion) of the reaction product obtained upon incubation of (28,28,28-2H3,28-13C)-24-methyldesmosterol with OsDWF1-expressing yeast (top panel) and vector control (bottom panel). The mass spectrum was recorded at the peak top of the product. The peak at tR = 17.74 min corresponds to the substrate. The data indicated that OsDWF1 was able to reduce the substrate to 24-methylcholesterol, although the enzyme activity was very poor.
Conversion of 24-Methylenecholesterol to 24-Methylcholesterol Using Transfected S2 Cells
It has been reported that the in vitro expression of A. thaliana DWF1 in yeast and Escherichia coli was not successful (11). The present failure of in vitro expression of OsDWF1 agreed with these previous results. Thus, we tried to express the OsDWF1 gene using an alternative system. S2 cells were transfected with OsDWF1, and the cells were incubated with (23,23,25-2H3,28-13C)-24-methylenecholesterol. GC-MS analysis, shown in Fig. 11, indicated that the exogenously added labeled substrate was converted to 24-methylcholesterol with a good yield. The mass spectrum of the resulting 24-methylcholesterol showed a molecular ion at m/z 475, which implies the loss of one deuterium atom, presumably from C-25, during the conversion. The data were consistent with that found for yeast in vivo. Therefore, it was concluded that OsDWF1 converts 24-methylenecholesterol to 24-methylcholesterol in the two-step mechanism.
FIGURE 11.
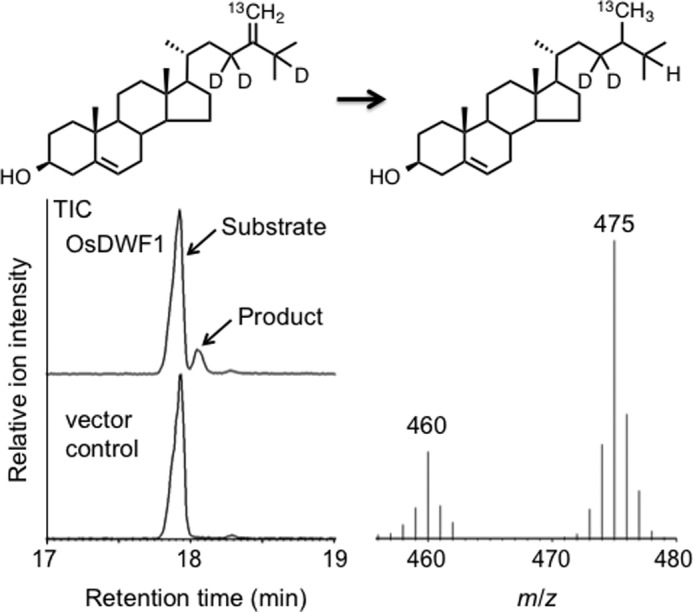
Shown is the GC-MS analysis of the reaction product obtained upon incubation of (23,23,25-2H3,28-13C)-24-methylenecholesterol with OsDWF1-transfected S2 cells (top panel) and the vector control (bottom panel). The MS spectrum was recorded at the peak top of the product, which clearly indicated that one of the deuterium atoms, presumably 25-2H, was eliminated during the conversion.
Discussion
This paper focused on a unique feature of ArDWF1. ArDWF1 was expressed in the yeast T21 strain. An in vivo assay indicated that ArDWF1 was able to convert endogenous 24-methylenecholetsterol to 24-methylcholesterol. In agreement with the in vivo data, 24-methylenecholetsterol was converted to 24-methylcholesterol in the in vitro assay. It was subsequently established that the 24-methylcholesterol produced consisted only of campesterol.
The mechanism of the ArDWF1-catalyzed reaction was investigated through labeling experiments. The C-25 hydrogen of 24-methylenecholesterol was retained during its conversion to campesterol in vivo and in vitro. These results were markedly different from those of typical biosyntheses of plant sterols in which the C-25 hydrogen was eliminated during the process. Indeed, a parallel experiment expressing OsDWF1 in yeast in vivo showed that the C-25 hydrogen was lost in the conversion of 24-methylenecholesterol to 24-methylcholesterol. The results presented here indicate that ArDWF1 catalyzes the direct reductive conversion of 24-methylenechol to campesterol without passing through the Δ24(25)-olefinic intermediate. This is the first report describing the direct reductive conversion of 24-methylenecholesterol to 24-methylcholesterol in higher plants as opposed to the two-step mechanism. 24-Methyldesmosterol was not converted to campesterol in the ArDWF1-expressing yeast in vitro, and this is understandable because the compound is not an obligatory intermediate in Ajuga plants.
Another interesting feature of ArDWF1 is that the formed 24-methylcholesterol consisted of solely campesterol. This is in contrast to the general view that 24-methylcholesterol is a C-24 epimeric mixture (campesterol and dihydrobrassicasterol) in higher plants. It has been reported that the 24-methylcholesterol of Glycine max (33), Zea mays (33), O. sativa (16), C. roseus (16), and A. thaliana (data not shown) consists of a mixture of campesterol and dihydrobrassicasterol. The composition of 24-methylcholesterol in the sterol fraction of Ajuga hairy roots was examined and determined to contain only campesterol, as revealed by chiral GC-MS analysis.
For comparative purposes, OsDWF1 was similarly expressed in yeast. 24-Methylenecholesterol was converted to 24-methylcholesterol (a mixture of campesterol and dihydrobrassicasterol) in an in vivo assay. The in vitro assay failed to reproduce the conversion, in contrast to the case of ArDWF1. However, OsDWF1 had a weak activity for the conversion of 24-methylenecholesterol to 24-methylcholesterol in vitro. The reason for its failure to convert 24-methylenecholesterol to 24-methylcholesterol remains unclear.
OsDWF1-transfected S2 cells were able to convert exogenously added 24-methylenecholesterol to 24-methylcholesterol with good efficiency. This finding suggests that S2 cells are an alternative host for DWF1 gene expression.
One may question whether a homolog of 24-methylenecholesterol, isofucosterol, can be a substrate of ArDWF1 and OsDWF1. Our preliminary results showed that isofucosterol was not converted to sitosterol with ArDWF1- and OsDWF1-expressing yeast T21 cells. Yeast T21 cells expressing the SSR2 genes of tomato and potato were reported to catalyze the reduction of desmosterol to cholesterol in vivo. Cholesterol is a minor sterol (approximately 5% of the total sterol of the Ajuga hairy roots). Therefore, it would be interesting to examine whether ArDWF1 converts desmosterol to cholesterol. The enzyme activities of ArDWF1 could be affected by the steroid nucleus structures of the substrates. A further systematic study including the incubation of various olefinic sterols having different nucleus structures are required to fully understand the substrate specificity of ArDWF1.
Historically, 24-methylcholesterol has been analyzed by GC and reported without defining the C-24 stereochemistry except for a few cases. At this time, the C-24 stereochemistry can be determined by high-resolution 1H NMR and chiral GC. The stereochemistry of C-24 will be a subject of further study to accumulate information on plants that contain solely campesterol as their 24-methylcholesterol. This will help answer how widely this unique DWF1 is distributed among higher plants. It remains a possibility that the unique sterol profile of Ajuga plants, which is devoid of sitosterol, is associated with the occurrence of this unique DWF1. Campesterol produced by ArDWF1, though a very minor sterol of Ajuga plants, may serve as a precursor of the plant hormone brassinolide (34).
The amino acid sequence of ArDWF1 showed a high similarity to those of known DWF1s, e.g. OsDWF1 (81% identity), A. thaliana DWF1 (79% identity), and G. max DWF1 (88% identity) (Fig. 2). Comparison of the amino acid sequence of ArDWF1 with those of known DWF1s revealed that there are more than 10 characteristic differences, including Ile460-Cys463, as shown in Fig. 2. To gain insights into which amino acid difference(s) is/are associated with the unique features of ArDWF1, site-directed mutagenesis studies are needed.
Δ24-Sterol reductases have been classified into two groups, Δ24(25)-sterol reductase (EC 1.3.1.72) and Δ24(28)-sterol reductase (EC 1.3.1.71). DHCR24 (Seladin-1) of human (35) and typical DWF1s are included in the former group, whereas the latter group includes ERG4 of yeast (Δ24(24(1))-sterol reductase) (36). ArDWF1 can be considered a member of the Δ24(28)-sterol reductase family on the basis of its substrate specificity and the reduction mechanism.
Author Contributions
Y. T., K. O., and Y. F. designed the study and wrote paper. Y. T. and K. O. characterized functions of DWF1s. H. Seki., T. A., T. M., and H. Suzuki designed and constructed the vectors for yeast expression. Y. F. synthesized the steroidal compounds. All authors analyzed the results and approved the final version of the manuscript.
Acknowledgments
We thank Drs. Takeshi Matsumoto (Daicel Co.) and Seiichi Toki (National Institute of Agrobiological Resources) for providing Ajuga hairy roots and O. sativa cell cultures, respectively. We thank Drs. Ryusuke Niwa (University of Tsukuba) for S2 cells and guidance regarding handling of the cells and Satoru Sawai (RIKEN Center for Sustainable Resource Science) for the yeast T21 strain.
This work was supported in part by Grant-in-Aid for Scientific Research (C) 26450138 (to K. O.) from the Ministry of Education, Culture, Sports, Science, and Technology. The authors declare that they have no conflicts of interest with the contents of this article.
The nucleotide sequence reported in this paper has been submitted to the DNA Data Bank of Japan with accession number LC070675 (ArDWF1).
- contig
- group of overlapping clones.
References
- 1.Nes W. R., and Mckean M. L. (1977) Biochemistry of Steroids and Other Isoprenoids, pp. 325–410, University Park Press, Baltimore [Google Scholar]
- 2.Patterson G. W., and Nes W. R. (1991) Physiology and Biochemistry of Sterols, pp. 83–117, American Oil Chemists' Society, Champain, IL [Google Scholar]
- 3.Schaller H. (2004) New aspects of sterol biosynthesis in growth and development of higher plants. Plant Physiol. Biochem. 42, 465–476 [DOI] [PubMed] [Google Scholar]
- 4.Lenton J. R., Goad L. J., and Goodwin T. W. (1975) Sitosterol biosynthesis in Hordeum vulgare. Phytochemistry 14, 1523–1528 [Google Scholar]
- 5.Uomori A., Nakagawa Y., Yoshimatsu S., Seo S., Sankawa U., and Takeda K. (1992) Biosynthesis of campesterol and dihydrobrassicasterol in cultured cells of Amsonia elliptica. Phytochemistry 31, 1569–1572 [Google Scholar]
- 6.Seo S., Uomori A., Yoshimura Y., Takeda K., Noguchi H., Ebizuka Y., Sankawa U., and Seto H. (1992) Biosynthesis of the 24-methylcholesterols dihydrobrassicasterol and campesterol in cultured cells of Amsonia elliptica: incorporation of [1,2-13C2]acetate and [2-13C,2H3]acetate. J. Chem. Soc. Perkin Trans. I 569–572 [Google Scholar]
- 7.Nes W. D., Song Z., Dennis A. L., Zhou W., Nam J., and Miller M. B. (2003) Biosynthesis of phytosterols: kinetic mechanism for the enzymatic C-methylation of sterols. J. Biol. Chem. 278, 34505–34516 [DOI] [PubMed] [Google Scholar]
- 8.Malhotra H. C., and Nes W. R. (1971) The mechanism of introduction of alkyl groups at C-24 of sterols IV: inhibition by triparanol. J. Biol. Chem. 246, 4934–4937 [PubMed] [Google Scholar]
- 9.Nes W. D., Janssen G. G., and Bergenstrahle A. (1991) Structural requirements for transformation of substrates by the (S)-adenosyl-l-methionine: Δ24(25)-sterol methyl transferase. J. Biol. Chem. 266, 15202–15212 [PubMed] [Google Scholar]
- 10.Fonteneau P., Hartmann-Bouillon M. A., and Benveniste P. (1977) A 24-methylene lophenol C-28 methyl transferase from suspension cultures of bramble cells. Plant. Sci. Lett. 10, 147–155 [Google Scholar]
- 11.Takahashi T., Gasch A., Nishizawa N., and Chua N. H. (1995) The DIMINUTO gene of Arabidopsis is involved in regulating cell elongation. Genes Dev. 9, 97–107 [DOI] [PubMed] [Google Scholar]
- 12.Klahre U., Noguchi T., Fujioka S., Takatsuto S., Yokota T., Nomura T., Yoshida S., and Chua N. H. (1998) The Arabidopsis DIMINUTO/DWARF1 gene encodes a protein involved in steroid synthesis. Plant Cell 10, 1677–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsumoto T., Wu J. Z., Kanamori H., Katayose Y., Fujisawa M., Namiki N., Mizuno H., Yamamoto K., Antonio B. A., Baba T., Sakata K., Nagamura Y., Aoki H., Arikawa K., Arita K., et al. (2005) Nature The map-based sequence of the rice genome. Nature 436, 793–800 [DOI] [PubMed] [Google Scholar]
- 14.Sawai S., Ohyama K., Yasumoto S., Seki H., Sakuma T., Yamamoto T., Takebayashi Y., Kojima M., Sakakibara H., Aoki T., Muranaka T., Saito K., and Umemoto N. (2014) Sterol side chain reductase 2 is a key enzyme in the biosynthesis of cholesterol, the common precursor of toxic steroidal glycoalkaloids. Plant Cell 26, 3763–3774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okuzumi T., Hara N., and Fujimoto Y. (1999) Mechanism of the double bond isomerization from Δ24(28) to Δ 24(25) in sitosterol biosynthesis in higher plants. Tetrahedron Lett. 40, 8863–8866 [Google Scholar]
- 16.Yamada J., Morisaki M., Iwai K., Hamada H., Sato N., and Fujimoto Y. (1997) 24-Methyl- and 24-ethyl-Δ24(25)-cholesterols as immediate biosynthetic precursors of 24-alkylsterols in higher plants. Tetrahedron 53, 877–884 [Google Scholar]
- 17.Fujimoto Y., Sato N., Iwai K., Hamada H., Yamada J., and Morisaki M. (1997) Stereochemistry of the reduction of 24-methyldesmosterol to campesterol and dihydrobrassicasterol in higher plants. Chem. Commun. 681–682 [Google Scholar]
- 18.Fujimoto Y., Sato N., Okuzumi T., Yamada J., and Morisaki M. (1998) Stereochemistry of the reduction of 24-ethyldesmosterol to sitosterol in tissue cultures of Oryza sativa. Bioorg. Med. Chem. Lett. 8, 205–208 [DOI] [PubMed] [Google Scholar]
- 19.Nagakari M., Kushiro T., Matsumoto T., Tanaka N., Kakinuma K., and Fujimoto Y. (1994) Incorporation of acetate and cholesterol into 20-hydroxyecdysone by hairy root clone of Ajuga reptans var. atropurpurea. Phytochemistry 36, 907–910 [Google Scholar]
- 20.Fujimoto Y., Ohyama K., Nomura K., Hyodo R., Takahashi K., Yamada J., and Morisaki M. (2000) Biosynthesis of sterols and ecdysteroids in Ajuga hairy roots. Lipids 35, 279–288 [DOI] [PubMed] [Google Scholar]
- 21.Hyodo R., and Fujimoto Y. (2000) Biosynthesis of 20-hydroxyecdysone in Ajuga hairy roots: the possibility of 7-ene introduction at a late stage. Phytochemistry 53, 733–737 [DOI] [PubMed] [Google Scholar]
- 22.Fujimoto Y., Maeda I., Ohyama K., Hikiba J., and Kataoka H. (2015) Biosynthesis of 20-hydroxyecdysone in plants: 3β-hydroxy-5β-cholestan-6-one as an intermediate immediately after cholesterol in Ajuga hairy roots. Phytochemistry 111, 59–64 [DOI] [PubMed] [Google Scholar]
- 23.Camps F., Coll J., and Cortel A., (1983) Sterols from Ajuga reptans (Labiatae). An. Quim. Ser. C. 79, 228–229 [Google Scholar]
- 24.Yagi T., Morisaki M., Kushiro T., Yoshida H., and Fujimoto Y. (1996) Biosynthesis of 24β-alkyl-Δ25-sterols in hairy roots of Ajuga reptans var. atropurpurea. Phytochemistry 41, 1057–1064 [Google Scholar]
- 25.Okuzumi K., Hara N., Fujimoto Y., Yamada J., Nakamura A., Takahashi K., and Morisaki M. (2003) Biosynthesis of phytoecdysteroids in Ajuga hairy roots: clerosterol as a precursor of cyasterone, isocyasterone and 29-norcyasterone. Tetrahedron Lett. 44, 323–326 [Google Scholar]
- 26.Tsukagoshi Y., Ohyama K., Seki H., Akashi T., Muranaka T., Suzuki H., and Fujimoto Y. (2016) Functional characterization of CYP71D443, a cytochrome P450 catalyzing C-22 hydroxylation in the 20-hydroxyecdysone biosynthesis of Ajuga hairy roots. [DOI] [PubMed] [Google Scholar]
- 27.Niwa R., Matsuda T., Yoshiyama T., Namiki T., Mita K., Fujimoto Y., and Kataoka H. (2004) CYP306A1, a cytochrome P450 enzyme, is essential for ecdysteroid biosynthesis in the prothoracic glands of Bombyx and Drosophila. J. Biol. Chem. 279, 35942–35949 [DOI] [PubMed] [Google Scholar]
- 28.Maruyama S., Fujimoto Y., Morisaki M., and Ikekawa N. (1982) Mechanism of campesterol demethylation in insect. Tetrahedron Lett. 23, 1701–1704 [Google Scholar]
- 29.Fujimoto Y., Ikuina Y., Nagakari M., Kakinuma K., and Ikekawa K. (1990) C-25 Prochirality in the fragmentation feaction catalysed by fucosterol epoxide lyase from the silkworm, Bombyx mori. J. Chem. Soc. Perkin Trans. I 2041–2046 [Google Scholar]
- 30.Takahashi K., Hashimoto K., Fujiyama A., Yamada J., Kobayashi N., Morisaki M., Nakano S., Hara N., and Fujimoto Y. (2003) Stereochemistry of reduction of the C-24,25 double bond in the conversion of desmosterol into cholesterol. Tetrahedron Lett. 44, 341–344 [Google Scholar]
- 31.Fujimoto Y., Kimura M., Khalifa F. A. M., and Ikekawa N. (1984) Side chain structural requirement for utilization of sterols by the silkworm for growth and development: non-stereoselective utilization of the 24-stereoisomeric pairs of 24-alkylsterols. Chem. Pharm. Bull. 32, 4372–4381 [Google Scholar]
- 32.Du L., and Poovaiah B. W. (2005) Ca2+/calmodulin is critical for brassinosteroid biosynthesis and plant growth. Nature 437, 741–745 [DOI] [PubMed] [Google Scholar]
- 33.Mulheirn L. J. (1973) Identification of C-24 alkylated steranes by P. M. R. spectroscopy. Tetrahedron Lett. 14, 3175–3178 [Google Scholar]
- 34.Grove M. D., Spencer G. F., Rohwedder W. K., Mandava N., Worley J. F., Warthen J. D. Jr., Steffens G. L., Flippen-Anderson J. L., and Cook J. C. Jr. (1979) Brassinolide, a plant growth-promoting steroid isolated from Brassica napus pollen. Nature 281, 216–217 [Google Scholar]
- 35.Waterham H. R., Koster J., Romeijn G. J., Hennekam R. C., Vreken P., Andersson H. C., FitzPatrick D. R., Kelley R. I., and Wanders R. J. (2001) Mutations in the 3β-hydroxysterol Δ24-reductase gene cause desmosterolosis, an autosomal recessive disorder of cholesterol biosynthesis. Am. J. Hum. Genet. 69, 685–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lai M. H., Bard M., Pierson C. A., Alexander J. F., Goebl M., Carter G. T., and Kirsch D. R. (1994) The identification of a gene family in the Saccharomyces cerevisiae ergosterol biosynthesis pathway. Gene 140, 41–49 [DOI] [PubMed] [Google Scholar]