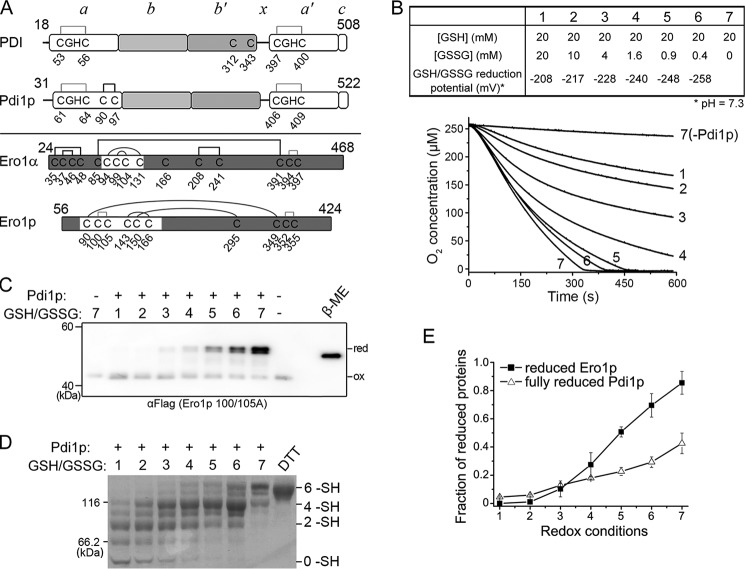
FIGURE 1.
Activation of Ero1p depends on fully reduced Pdi1p. A, upper, cysteine connectivities of oxidized PDI and Pdi1p were shown schematically with the -CGHC- active sites in gray lines and structural disulfide in black line. Lower, cysteine connectivities of fully oxidized Ero1α and Ero1p were shown schematically with the disulfides of active sites in gray lines, regulatory disulfides in black curves, and structural disulfides in black lines. The starting and ending residue numbers of protein constructs were labeled on the upper portion of each diagram. B, upper, the glutathione redox buffers with different reduction potentials used in the oxygen consumption assay. The reduction potential was calculated according to the Nernst equation (Eh′ = E0′ − RT/nF × ln([GSH]2/[GSSG]), where E0′ = −258 mV, pH 7.3 (37), and n = 2). Lower, the oxygen consumption catalyzed by 2 μm Ero1p during oxidation of 20 μm Pdi1p in each glutathione redox buffer. C, enzymatically inactive Ero1p C100A/C105A-FLAG at 0.5 μm was incubated with or without 10 μm Pdi1p in corresponding glutathione redox buffer as numbered in B for 30 min. The oxidized (ox) and reduced (red) forms of Ero1p were monitored after NEM blocking by Western blotting of non-reducing SDS-9% PAGE using αFLAG. Fully reduced Ero1p was prepared with excess β-mercaptoethanol (β-ME). Note that the slow-migrating doublet of Ero1p is due to the heterogeneous reduction of Cys90-Cys349 and Cys150-Cys295 long-range disulfides. D, in the same reaction as in C, the redox states of Pdi1p were monitored by Coomassie staining after mPEG-5k modification; Pdi1p reduced by excess DTT was loaded as a marker for fully reduced Pdi1p. The number of free thiols (-SH) in each Pdi1p species was labeled on the right margin. Note that each of the 6 -SH, 4 -SH, and 2 -SH species was accompanied with a faster migrating band, probably due to incomplete alkylation. E, the fraction of reduced Ero1p doublet in C and fully reduced Pdi1p in D was quantified by densitometry and plotted against the redox conditions in B (mean ± S.D., n = 3 independent experiments), respectively.