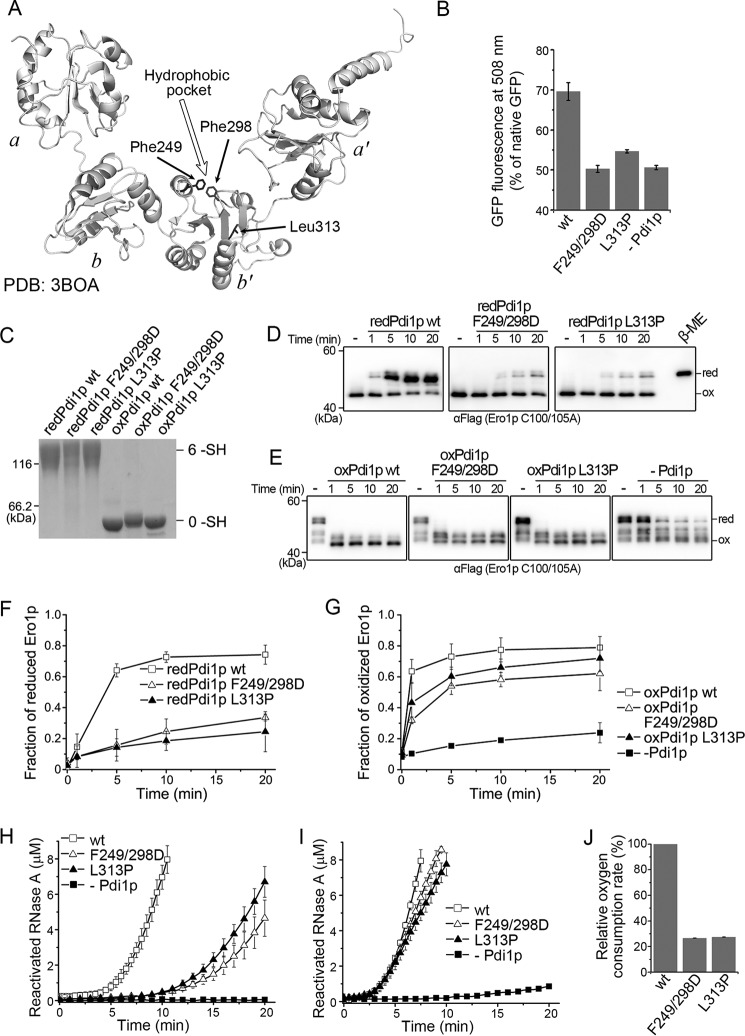
FIGURE 6.
Effects of client-binding mutations in Pdi1p b′ domain on Ero1p-Pdi1p interplay. A, ribbon diagram of Pdi1p displaying the locations of Phe249, Phe298, and Leu313. B, the refolding yield of 50 nm acid-denatured green fluorescent protein was determined by fluorescence intensity in the absence or presence of 1 μm Pdi1p proteins as indicated. The fluorescence intensity of 50 nm native GFP was taken as 100% (mean ± S.D., n = 3 independent experiments). C, the redox states of reduced and oxidized Pdi1p client-binding mutants were monitored as described in the legend to Fig. 2A. D, the reduction of the regulatory disulfides in Ero1p C100A/C105A-FLAG by reduced Pdi1p substrate binding mutants was monitored as described in the legend to Fig. 3B. E, the re-oxidation of the regulatory disulfides in Ero1p C100A/C105A-FLAG by oxidized Pdi1p substrate-binding mutants was monitored as described in the legend to Fig. 3C. F and G, the fraction of reduced Ero1p doublet in D and oxidized Ero1p species in E was quantified by densitometry and plotted against time (mean ± S.D., n = 3 independent experiments), respectively. H and I, reactivation of 8 μm reduced and denatured RNase A by 3 μm Pdi1p proteins as indicated in the presence of 3 μm hyperactive Ero1p C150A/C295A (H) or 1 mm GSH and 0.2 mm GSSG (I) was determined (mean ± S.D., n = 3 independent experiments). J, the oxygen consumption catalyzed by 2 μm hyperactive Ero1p C150A/C295A during the oxidation of 20 μm Pdi1p in 10 mm GSH was determined as described in the legend to Fig. 5A (mean ± S.D., n = 3 independent experiments).