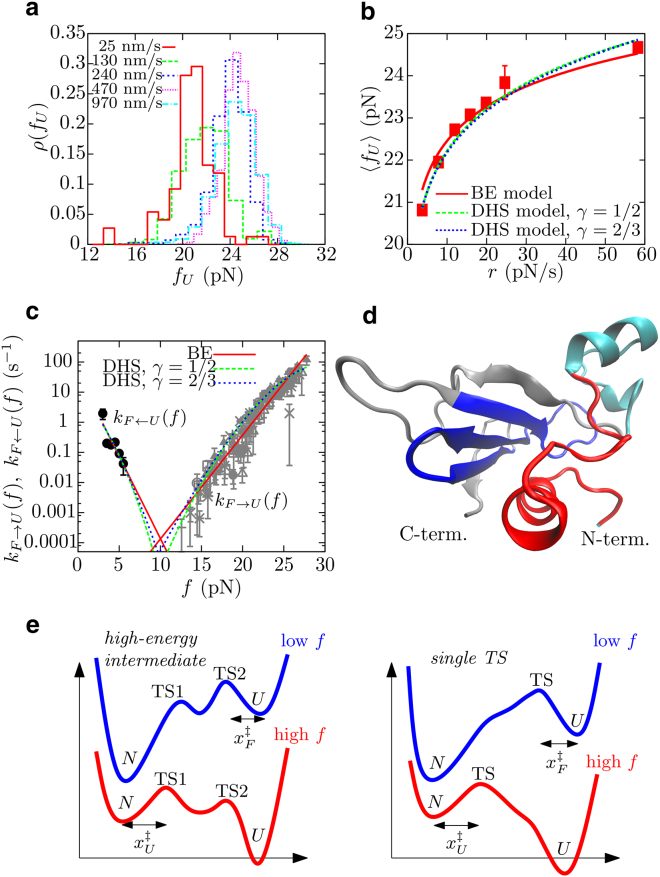
Figure 3.
Mechanical unfolding, folding, and TS of barnase. (a) Unfolding force histograms obtained at different pulling speeds. (b) Dependence of average unfolding forces 〈fU〉 with loading rate r (r = , where ∼ 0.069 pN/nm). Fits to the analytical expressions provided by the BE (red line; Eq. S6) and DHS (green and blue lines for γ = 1/2 and 2/3, respectively; Eq. S8) models are shown. (c) Unfolding and folding kinetic rates, kF→U(f) (gray, each symbol is associated to a different pulling speed in the range 25–960 nm/s) and kF←U(f) (black), respectively, as a function of force. Fits to the analytical expressions provided by the BE and DHS models are shown (Eqs. S5 and S7, respectively). Color code as in (b). (d) Structure of barnase in the folded state. (Red) The first 25 amino acids to unfold in the mechanical unfolding process; (cyan) helices 2 and 3, which first unfold in chemical denaturation. (Blue) The hydrophobic core of the protein, dominated by β-sheets (made of a total of 29 amino acids); its formation corresponds to the TS-mediating mechanical folding from state U. (e) Two possible scenarios explain our experimental results for the mFEL of the protein barnase. (Left) There is a high-energy intermediate state surrounded by TS1, that is located close to state N and mediates unfolding at high forces, and TS2, that is located close to state U and mediates folding at low forces. (Right) The mFEL has a single TS that changes its position along the reaction coordinate as a function of force. To see this figure in color, go online.