Precise genome editing enables an herbicide tolerance trait in a commercially relevant crop by improving the outcome of DNA double strand break repair using single-stranded oligonucleotides.
Abstract
Here, we report a form of oligonucleotide-directed mutagenesis for precision genome editing in plants that uses single-stranded oligonucleotides (ssODNs) to precisely and efficiently generate genome edits at DNA strand lesions made by DNA double strand break reagents. Employing a transgene model in Arabidopsis (Arabidopsis thaliana), we obtained a high frequency of precise targeted genome edits when ssODNs were introduced into protoplasts that were pretreated with the glycopeptide antibiotic phleomycin, a nonspecific DNA double strand breaker. Simultaneous delivery of ssODN and a site-specific DNA double strand breaker, either transcription activator-like effector nucleases (TALENs) or clustered, regularly interspaced, short palindromic repeats (CRISPR/Cas9), resulted in a much greater targeted genome-editing frequency compared with treatment with DNA double strand-breaking reagents alone. Using this site-specific approach, we applied the combination of ssODN and CRISPR/Cas9 to develop an herbicide tolerance trait in flax (Linum usitatissimum) by precisely editing the 5′-ENOLPYRUVYLSHIKIMATE-3-PHOSPHATE SYNTHASE (EPSPS) genes. EPSPS edits occurred at sufficient frequency that we could regenerate whole plants from edited protoplasts without employing selection. These plants were subsequently determined to be tolerant to the herbicide glyphosate in greenhouse spray tests. Progeny (C1) of these plants showed the expected Mendelian segregation of EPSPS edits. Our findings show the enormous potential of using a genome-editing platform for precise, reliable trait development in crop plants.
With humanity facing significant challenges to increase global agricultural productivity, there is an urgent need for trait development and improvement. Novel methods for targeted editing of plant genomic DNA will play an important role in addressing these challenges. Oligonucleotide-directed mutagenesis (ODM) is a nontransgenic base pair-specific precision genome editing platform that has been employed successfully in bacterial, fungal, mammalian, and plant systems (Moerschell et al., 1988; Yoon et al., 1996; Beetham et al., 1999; Zhu et al., 1999; Aarts et al., 2006; Gocal et al., 2015; Sauer et al., 2016). ODM employs chemically synthesized oligonucleotides to mediate genome editing by acting as DNA templates during the editing process. In the early application of this technology in plants, oligonucleotides were used to target the acetolactate synthase (ALS) gene in tobacco (Nicotiana tabacum; Beetham et al., 1999), in maize (Zea mays; Zhu et al., 1999), and later in oilseed rape (Brassica napus; Gocal et al., 2015). In both tobacco and maize, a DNA/RNA chimeric oligonucleotide was used to target the ALS gene, whereas for oilseed rape, an oligonucleotide containing a 5′Cy3 dye and a 3′idC reverse base was employed. These terminal chemistries prevent the oligonucleotide from undergoing recombination but still allow it to act as a mutagen and DNA template for gene editing. In many peer-reviewed ODM studies, one of the most frequently used chemical modifications for oligonucleotides is phosphorothioate linkages on the terminal bases (Andrieu-Soler et al., 2005; Radecke et al., 2006; de Piédoue et al., 2007). In mammalian cells, oligonucleotide toxicity is correlated with the number of terminal phosphorothioate linkages as measured by fractional cell survival or γ-H2AX phosphorylation (Olsen et al., 2005; Rios et al., 2012). In plants, oligonucleotides containing a 5′Cy3 dye and a 3′idC reverse base chemistry proved to be less toxic than oligonucleotides with terminal phosphorothioate linkages in most crop varieties tested when measured in cell survival assays (Schöpke CR, Segmi RE, Narváez-Vásquez J, unpublished data).
Genome editing using reagents that cause double strand breaks (DSBs) in either a nontargeted or targeted manner is enhanced by ODM (Menke et al., 2001; Suzuki et al., 2003; Strouse et al., 2014; Bialk et al., 2015). These reagents include the bleomycin family of glycopeptide antibiotics and, more recently, engineered nucleases such as meganucleases, zinc finger nucleases, transcription activator-like effector nucleases (TALENs), and clustered, regularly interspaced, short palindromic repeats (CRISPR/Cas9). Gene targeting in mammalian cells was increased when oligonucleotides were used along with a low dose of the nontargeted DSB reagent bleomycin (Suzuki et al., 2003). This effect is hypothesized to result from increased expression/activity of DNA repair genes and/or in the presence of DSBs near the target site. However at high doses, glycopeptide antibiotics are particularly toxic to dividing cells, thereby constraining their use as tools for genome editing. Ideally, the generation of a single DSB in close proximity to the desired genomic target could increase the editing efficiency while at the same time obviating the genotoxic effect of chemical reagents like glycopeptide antibiotics. This has been achieved with the recent advent of engineered nucleases, where target-specific DSBs are now routine.
In plants, DSBs are typically repaired by the imprecise nonhomologous end joining (NHEJ) DNA repair pathway, resulting in random deletions and insertions (indels) at the site of repair (Schröpfer et al., 2014). By leveraging the error-prone nature of NHEJ, many groups have reported using engineered nucleases to generate loss-of-function alleles in a target-specific manner (Carlson et al., 2012; Lor et al., 2014; Shan et al., 2015; Zhang et al., 2016). Precision genome editing, on the other hand, relies on template-directed repair of DSBs using exogenously supplied double-stranded DNA or single-stranded oligonucleotides (ssODNs) and, as such, is more precise than NHEJ (Voytas, 2013). While groups have reported using oligonucleotides in combination with engineered nucleases to increase gene-editing frequencies in fish, mammals, and flies, in plants, the challenge of making and tracking targeted genome edits from individual cells to whole plants in a nonselectable manner remains a barrier.
Here, we report a significant improvement in genome editing when ssODNs are used to reliably and precisely target nucleotide sequence changes close to a cut site made by DSBs in either a nontargeted or targeted manner. Importantly, we show that these precise, scarless edits are made at sufficient frequency that they can be tracked from individual cells to whole plants without the use of selection. This nontransgenic approach to plant genome editing allows for the development of novel traits, both selectable and nonselectable, in agriculturally important crop species.
RESULTS
ssODNs Combined with Glycopeptide Antibiotic
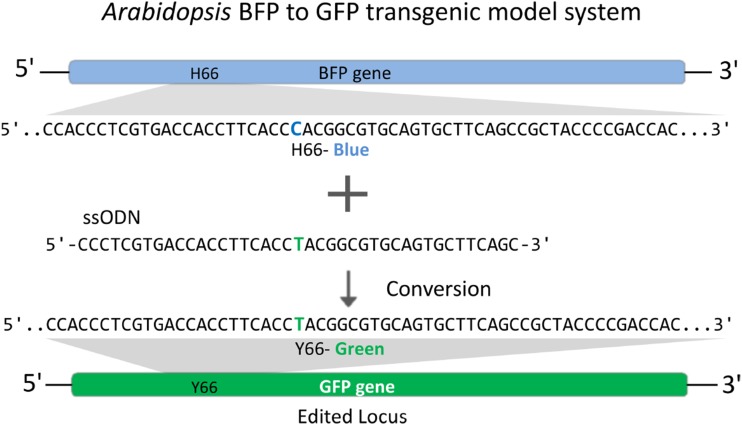
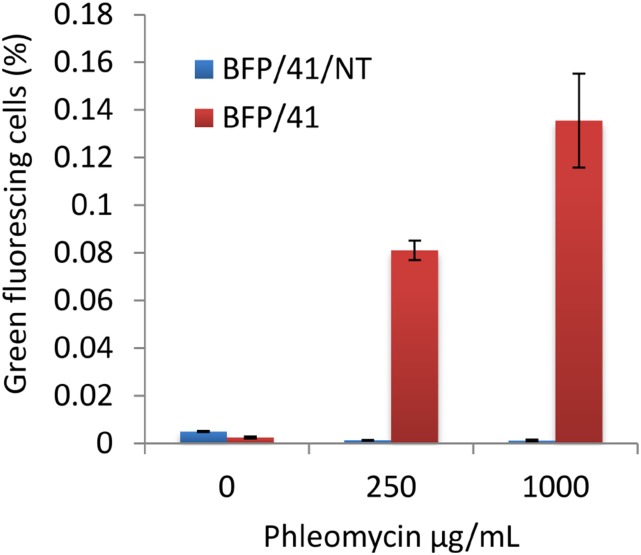
To test the effect of ssODN in combination with low-dose glycopeptide antibiotic treatment, we used an Arabidopsis (Arabidopsis thaliana) transgenic line in which a stably integrated blue fluorescent protein gene (BFP) can be converted to a GFP gene by editing the codon His-66 (CAC) to Tyr-66 (TAC; Fig. 1). Using this system, we can quantify genome editing based on a cell’s green fluorescence using flow cytometry. Protoplasts from this line were treated for 90 min with 0, 250, or 1,000 µg mL−1 of the glycopeptide antibiotic phleomycin. We then introduced either ssODN BFP/41 or BFP/41/NT (BFP/41/NT serves as a negative control and does not contain the C→T edit to convert BFP to GFP; Supplemental Table S1) and monitored GFP fluorescence by cytometry 24 h after delivery. BFP/41 along with phleomycin pretreatment resulted in a dose-dependent increase in the number of GFP-positive cells (Fig. 2). These results provide evidence that ssODNs can enhance the frequency and precision of nonspecific DSB reagents, such as phleomycin-based genome editing in Arabidopsis protoplasts.
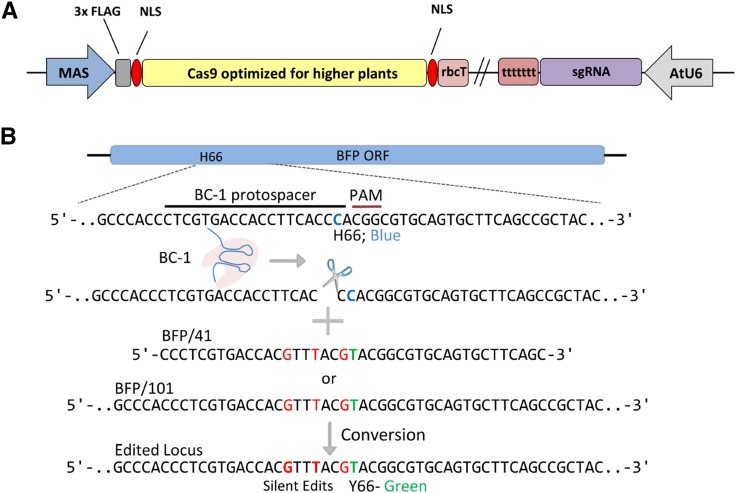
Figure 1.
Diagram of the Arabidopsis BFP-to-GFP transgenic system for detecting ssODN-mediated precision genome editing. The edited nucleotide change (C→T) that results in the conversion from BFP to GFP fluorescence is shown as blue and green letters, respectively.
Figure 2.
Effects of ssODNs on BFP-to-GFP editing in Arabidopsis protoplasts pretreated with phleomycin. Protoplasts were pretreated with 0, 250, or 1,000 µg mL−1 phleomycin for 90 min prior to the delivery of ssODN BFP/41 or BFP/41/NT. GFP fluorescence was measured by cytometry 24 h after the delivery of ssODNs. Data represent means ± se (n = 4).
ssODNs Combined with TALEN
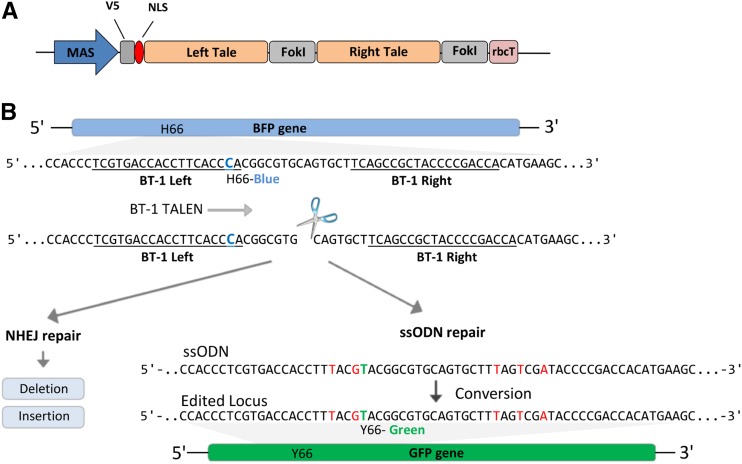
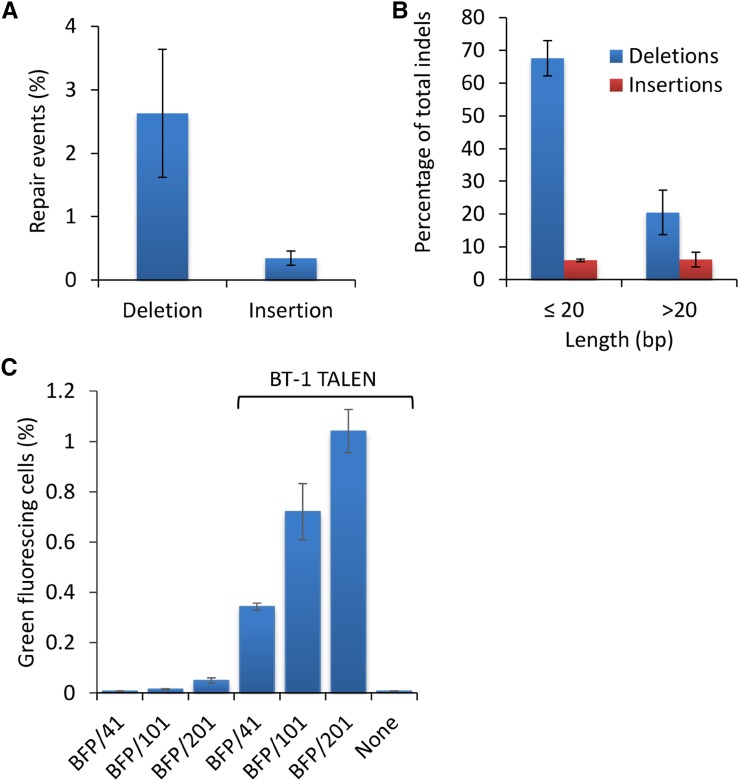
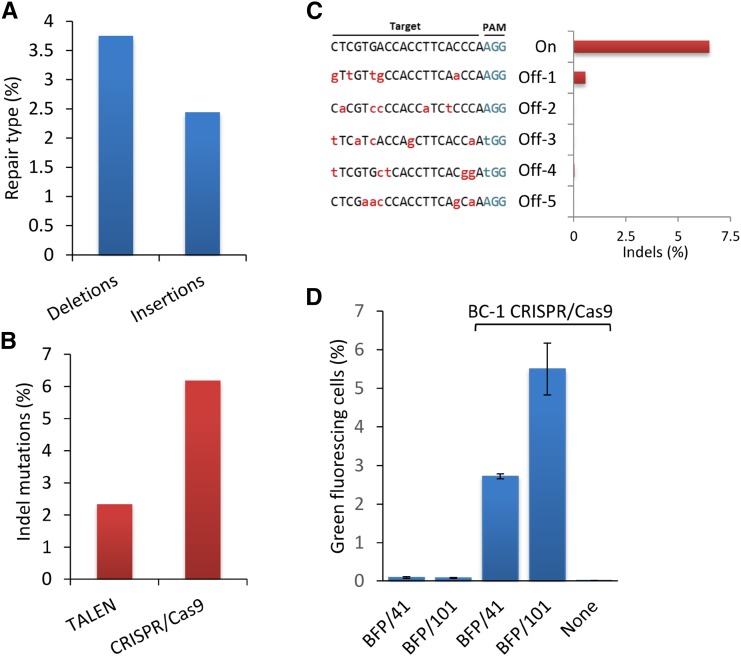
We next asked if ssODNs can also positively influence genome-editing outcomes induced by DSB reagents that make a more target-specific approach. To test this, we engineered a TALEN expression construct (BT-1; Fig. 3A) that will target and make a DSB just downstream of the C→T edit required to convert BFP to GFP in our Arabidopsis transgenic line (Fig. 3B). The BT-1 TALEN construct consists of two arms, both having a TAL effector-like DNA-binding domain (TALE) linked to a catalytic DNA nuclease domain of FokI. The TALE domains guide the TALEN arms to specific sites of DNA, allowing for dimerization of the FokI endonucleases and subsequent generation of a targeted DNA DSB in the spacer region between the two binding sites (Cermak et al., 2011). Each BT-1 TALE recognizes a 19-bp sequence separated by a 14-bp spacer and is composed of the truncated N152/C+63 architecture (Miller et al., 2011; Supplemental Table S2). We first examined BT-1 activity at the targeted site on the BFP transgene by measuring imprecise NHEJ repair events occurring in the spacer region. To accomplish this, total genomic DNA was extracted from treated protoplasts 72 h after the introduction of BT-1 and the target region amplified by PCR. PCR amplicons were then deep sequenced to a depth of greater than 500,000 reads (Supplemental Fig. S1A; Supplemental Table S3). Analysis showed that the frequency of deletions and insertions averaged 2.6% and 0.3%, respectively (Fig. 4A). Deletions were primarily 20 bp or less, while insertions were more equally distributed with respect to size (Fig. 4B). After establishing the targeting activity of BT-1 on the BFP transgene, we next tested the effect of combining ssODNs with BT-1 TALEN to convert BFP to GFP. For these experiments, we examined ssODNs of three different lengths (BFP/41, BFP/101, and BFP/201; Supplemental Table S1), each independently delivered with or without BT-1 plasmid into Arabidopsis protoplasts. The resulting BFP-to-GFP editing was then quantified by cytometry 72 h after delivery. Protoplasts treated with both ssODNs and BT-1 TALEN exhibited 25- to 45-fold more green-fluorescing cells than treatment with ssODN alone and more than 125-fold when TALENs were used alone (Fig. 4C). Notably, ssODN length had a positive effect on the frequency of BFP-to-GFP edits, whether combined with BT-1 TALEN or used alone. Taken together, these data show that when ssODNs are combined with a target-specific DSB reagent, the frequency of precise genome edits is increased by approximately 10-fold over that observed with a nonspecific DSB reagent.
Figure 3.
BT-1 TALEN design and target diagram. A, The mannopine synthase (MAS) promoter drives the expression of the TALEN monomers. The pea (Pisum sativum) rbcT RBCSE9 acts as a gene terminator. A V5 epitope tag and an SV40 nuclear localization signal (NLS) reside on the N terminus. B, BFP target region schematic. The BT-1 TALE left and right binding domains are underlined. The site of BFP-to-GFP conversion (C→T) is in blue. BT-1 activity produces a DSB that can be repaired by NHEJ or through ssODNs, resulting in indels or BFP-to-GFP precision editing, respectively. Red bases are silent substitutions used to discourage BT-1 activity after conversion.
Figure 4.
Addition of ssODNs enhances genome editing in Arabidopsis protoplasts treated with TALEN BT-1. A, Imprecise NHEJ repair events in Arabidopsis protoplasts treated with BT-1 TALEN. B, Percentage of total indels as a result of BT-1 activity by length. C, Addition of ssODN to BT-1 TALEN-treated protoplasts significantly enhances the frequency of BFP-to-GFP conversion in Arabidopsis. Arabidopsis protoplasts were treated with BFP/41, BFP/101, or BFP/201 with and without BT-1 TALEN or BT-1 TALEN without ssODN. BFP-to-GFP edits were evaluated by cytometry 72 h after delivery. Data represent means ± se (n = 3).
ssODNs Combined with CRISPR/Cas9
Combining ssODNs with TALENs resulted in a significant improvement in the frequency of genome edits over using TALENs or ssODNs alone. However, considering the complexity of reengineering TALEN proteins for each new DNA target, we asked if the more easily designed and constructed engineered nuclease CRISPR/Cas9 also could show enhanced genome-editing frequency when supplied with ssODNs. The CRISPR/Cas9 system consists of a Streptococcus pyogenes Cas9 nuclease and a chimeric fusion of two RNAs (crRNA and tracrRNA) referred to as an engineered single guide RNA (sgRNA). The sgRNA supports targeted nucleic acid specificity for Cas9 through base pairing of its first 20 5′ bases with the DNA target, resulting in a site-specific DSB (Cong et al., 2013). In contrast with TALENs, changing the target specificity of the CRISPR/Cas9 protein complex does not require extensive protein engineering but only minimal manipulation of the sgRNA. We designed the CRISPR/Cas9 expression plasmid, BC-1 (Fig. 5A), to target near locus His-66 of the BFP gene in our transgenic model (Fig. 5B; Supplemental Table S4). Following a similar experimental methodology to that in our TALEN work, we first examined the ability of BC-1 to target and cleave the BFP gene by measuring the frequency of imprecise NHEJ repair events found upstream of the protospacer adjacent motif (PAM) sequence. In protoplasts treated with BC-1, we detected deletions and insertions at a frequency of 3.7% and 2.4%, respectively, using deep amplicon sequencing (Fig. 6A). The most represented indel for either insertions or deletions was a single base pair (data not shown). Notably, when compared with similar experiments with BT-1 (TALEN), the BFP transgene-targeting efficiency of BC-1 (CRISPR/Cas9) was nearly 3 times higher (Fig. 6B). Having established the on-target activity of BC-1, we next tested for potential off-target cleavage by searching the Arabidopsis genome for sequences with high similarity to the BC-1 target sequence using Cas-OFFinder (Bae et al., 2014). We examined five potential off-target sites that, based on searches, exhibited the most homology to the BC-1 target sequence (Hsu et al., 2013). Arabidopsis protoplasts were treated with BC-1 for 72 h, after which amplicons were generated using primers that flank each of the five potential off-target sites (Supplemental Table S3). These amplicons were then analyzed for NHEJ mutations by amplicon deep sequencing. Of the five sites tested, only Off-1 showed mutations near the predicted cleavage site (Fig. 6C; Supplemental Table S5). While detectable, this level is approximately 13-fold less than the on-target control. We suspect that this weak activity at Off-1 is based on the homology of the sequence proximal to the PAM site where only one mismatch is present (Fig. 6C; Hsu et al., 2013).
Figure 5.
CRISPR/Cas9 construct design and target region diagram. A, The MAS promoter drives the transcription of the plant codon-optimized SpCas9 gene that contains two SV40 NLSs at the N and C termini and a 3× FLAG epitope tag on the N terminus. The pea rbcT RBCSE9 acts as a gene terminator. The AtU6-26 promoter drives the transcription of the poly(T) terminated sgRNA scaffold. B, Approach used to target the BFP transgene using BC-1 CRISPR/Cas9. The protospacer is shown as a black line and the protospacer adjacent motif (PAM) as a red line. The edited nucleotide change (C→T) resulting in the conversion from BFP to GFP is shown as blue and green letters, respectively. Red nucleotides are silent mutations used to deter BC-1 activity on a converted GFP transgene.
Figure 6.
BC-1 CRISPR/Cas9 activity in Arabidopsis protoplasts. A, Imprecise NHEJ repair events in protoplasts treated with BC-1 as determined by amplicon deep sequencing (n = 1). B, Activity of TALEN BT-1 contrasted with CRISPR/Cas9 BC-1 in Arabidopsis protoplasts as determined by the percentage of imprecise NHEJ events. C, Off-target analysis for BC-1 CRISPR/Cas9. Imprecise NHEJ events at five loci homologous to the BC-1 target sequence were measured by amplicon deep sequencing (n = 1). Bases in lowercase red letters are mismatches to the BC-1 target sequence. D, ssODNs enhance BFP-to-GFP editing in Arabidopsis protoplasts treated with BC-1. Protoplasts were treated with BFP/41 or BFP/101 with and without CRISPR/Cas9 or BC-1 CRISPR/Cas9 alone. BFP-to-GFP edits were measured by flow cytometry 72 h after delivery. Data represent means ± se (n = 5).
Next, to examine the ability of ssODNs to enhance BC-1-mediated BFP-to-GFP editing, we delivered either ssODN BFP4/41 or BFP4/101 along with BC-1 into protoplasts and then measured GFP fluorescence 72 h later. When BFP/41 or BFP/101 was delivered with BC-1 to protoplasts, a marked increase in the number of GFP positive cells was observed compared with treatments without ssODNs (Fig. 6D). Analogous to our findings with TALENs, we observed a similar ssODN length-dependent effect on editing frequency with CRISPR/Cas9. In comparison, ssODNs added to CRISPR/Cas9 introduced to protoplasts resulted in nearly 8 times more BFP-to-GFP edits than in experiments using TALENs, a result that we suspect is based partially on the higher targeting activity observed with BC-1 on the BFP transgene. Collectively, these results demonstrate that BC-1 can actively target and disrupt the BFP transgene and leave negligible off-target footprints. Moreover, when precise cuts made by BC-1 are corrected using ssODNs, the frequency of precise and scarless BFP-to-GFP edits in Arabidopsis protoplasts is greater compared with when BC-1 or ssODNs are used alone.
Establishing Precise 5′-ENOLPYRUVYLSHIKIMATE-3-PHOSPHATE SYNTHASE Gene Edits in Flax Using ssODNs and CRISPR/Cas9
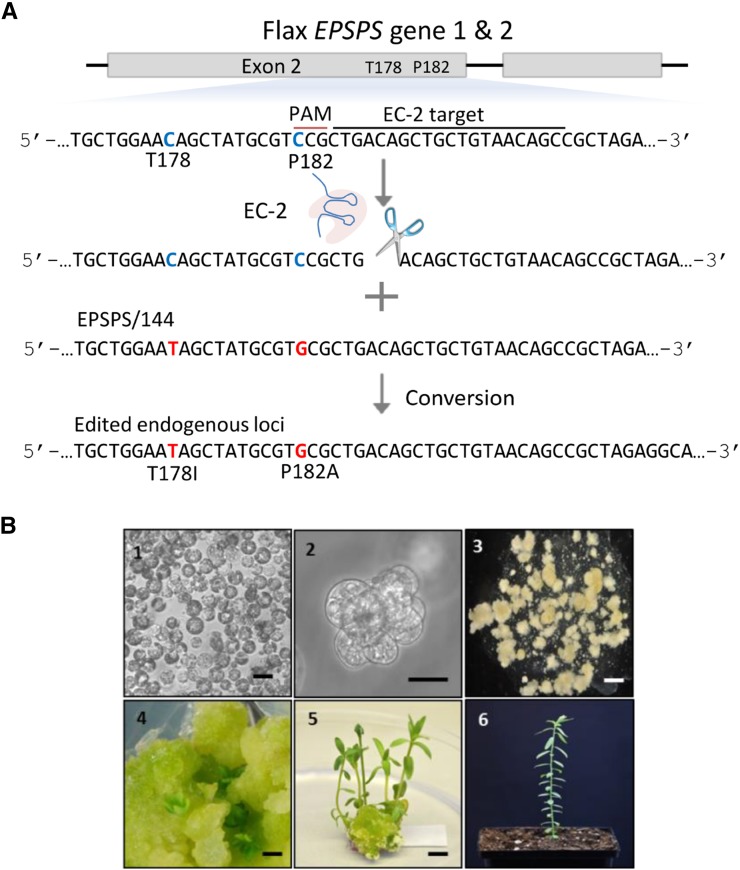
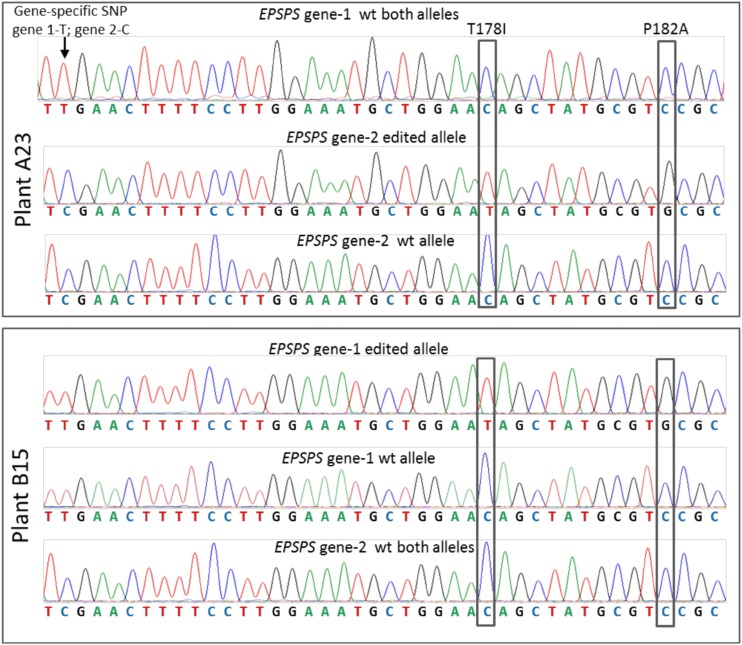
To extend the application of genome editing using ssODNs combined with an engineered nuclease to a commercially relevant agricultural crop, we conducted a series of experiments targeting the two highly homologous 5′-ENOLPYRUVYLSHIKIMATE-3-PHOSPHATE SYNTHASE (EPSPS) genes in flax (Linum usitatissimum; Supplemental Text S1). The EPSPS genes code for a protein in the shikimate pathway that participates in the biosynthesis of aromatic amino acids. In plants, EPSPS is a target for glyphosate, an herbicide that acts as a competitive inhibitor of the binding site for phosphoenolpyruvate (Schönbrunn et al., 2001). Based on our work in Arabidopsis, we chose to make precise edits in the flax EPSPS genes, using ssODNs combined with CRISPR/Cas9 components. We designed a CRISPR/Cas9 expression plasmid (EC-2) that targets a conserved sequence in both EPSPS genes near two loci, Thr-178 and Pro-182, that, when edited to Ile-178 and Ala-182, will render the EPSPS enzyme tolerant to glyphosate (Gocal et al., 2007; Fig. 7A; Supplemental Table S4). We then delivered the ssODN EPSPS/144 containing the two targeted changes, one of which will disrupt the PAM sequence (Fig. 7A; Supplemental Table S1), together with EC-2 into flax protoplasts. The treated protoplasts were then allowed to divide to form microcolonies without using selection for 21 d (Fig. 7B). We then measured for precise edits and indel scars in both EPSPS loci by PCR amplifying the region surrounding the target site and subjecting the amplicons to deep sequencing (Supplemental Fig. S1C). The frequency of precise EPSPS edits ranged between 0.09% and 0.23%, and indels between 19.2% and 19.8%, in three independent experiments with these edits and indels being equally distributed between the two loci (Table I). After establishing the presence of T178I and P182A edits in microcolonies, we next regenerated calli, again without employing any selective agent, then molecularly screened for the targeted edits and indel scars using allele-specific quantitative PCR (Morlan et al., 2009). Of 5,167 calli screened from experiment 1 and 4,601 from experiment 2, eight (0.15%) and four (0.08%) contained both T178I and P182A changes in at least one of the EPSPS loci, respectively (Table I). This edit frequency correlated with the initial sequencing of 21-d-old microcolonies (Table I). Calli that screened positive for precise edits from experiment 1 were used to regenerate whole plants under nonselective conditions, 100% of which screened positive for the presence of the T178I and P182A edits in at least one EPSPS gene through DNA cloning and Sanger sequencing. Sequencing traces for two of these EPSPS edited plants, A23 and B15, are shown in Figure 8. All regenerated plants transferred to soil were fertile and genotyped as heterozygous for the T178I and P182A edits at either the gene 1 or gene 2 locus. No plants were biallelic or heterozygous for both genes. C1 progeny from several A23 line plants derived from a single callus event were then evaluated for inheritance of the edited EPSPS allele. Sequence analysis showed sexual transmission of the edited EPSPS allele with the expected Mendelian segregation ratio of 1:2:1 (Table II).
Figure 7.
Approach used to target the EPSPS loci in flax. A, Target region of the EPSPS loci Thr-178 and Pro-182 of exon 2. The EC-2 protospacer is shown as a black line, and the PAM is shown as a red line. The nucleotides within the codons targeted for edit are in blue (ACA and CCG), and edited nucleotides are in red (ATA and GCG). The CCG→GCG edit disrupts the PAM, minimizing EC-2 activity on an edited gene. B, Stages in the flax genome-editing workflow. Image 1, Protoplasts (bar = 10 µm); image 2, microcolony at 3 weeks (bar = 50 µm); image 3, microcalli at 7 weeks (bar = 100 µm); image 4, shoot initiation from callus (bar = 0.5 cm); image 5, regenerated shoots (bar = 0.5 cm); and image 6, regenerated plant in soil.
Table I. Summary of flax CRISPR/Cas9 experiments targeting EPSPS.
| Experiment | Deep Sequencing of Microcoloniesa |
Calli Genotyping Resultsb |
||
|---|---|---|---|---|
| Precise Editsc | Indels | Calli Screened | Calli with Precise Edits | |
| FC-1 | 0.23% | 19.8 | 5,167 | 8 (0.15%) |
| FC-2 | 0.10% | 19.2 | 4,601 | 4 (0.08%) |
| FC-3 | 0.09% | 19.6 | NS | |
NS, Experiment was not screened.
Genomic DNA was isolated from pools of approximately 10,000 microcolonies and then used as a template to amplify the target region.
bIndividual callus was screened first by allele-specific PCR and then confirmed by Sanger sequencing.
cSequences with T97I (ACA→ATA) and P101A (CCG→GCG); data were combined for gene 1 and gene 2.
Figure 8.
Sequence confirmation of the edited EPSPS alleles in regenerated plants A23 and B15. The arrow shows a gene-specific single-nucleotide polymorphism (SNP). The boxed areas show the T178I and P182A edits. Regenerated plant A23 contains the T178I and P182A precise edits in one allele of EPSPS gene 2. Regenerated plant B15 contains the T178I and P182A precise edits in one allele of EPSPS gene 1. Chromatograms are representative of multiple genomic DNA extractions from each plant. wt, Wild type.
Table II. Transmission of targeted EPSPS edits to the progeny of selfed C0 plants (C1 generation).
| C0 Plant No. | Genotype of EPSPS Gene 2 |
|||
|---|---|---|---|---|
| Wild Type | Heterozygous for Ile-178 and Ala-182 | Homozygous for Ile-178 and Ala-182 | χ2a | |
| 1 | 19 | 31 | 21 | 1.25 |
| 2 | 15 | 29 | 20 | 1.34 |
| 3 | 4 | 11 | 2 | 1.94 |
The observed ratio is not significantly different from expected for all C1 progeny.
χ2 test with 2 degrees of freedom at P = 0.05; expected ratio of 1:2:1.
To identify potential off-target mutations arising from treatment with EC-2 in regenerated plant A23, we amplified eight different regions of the flax genome bearing sequence similarity to the EC-2 protospacer. We then measured for NHEJ mutations made through imprecise NHEJ events by amplicon deep sequencing. Mutations indicative of EC-2 activity were not detected in any of the eight potential off-target sites tested for plant A23 (Supplemental Table S6).
Glyphosate Tolerance of Edited Callus and Whole Plants
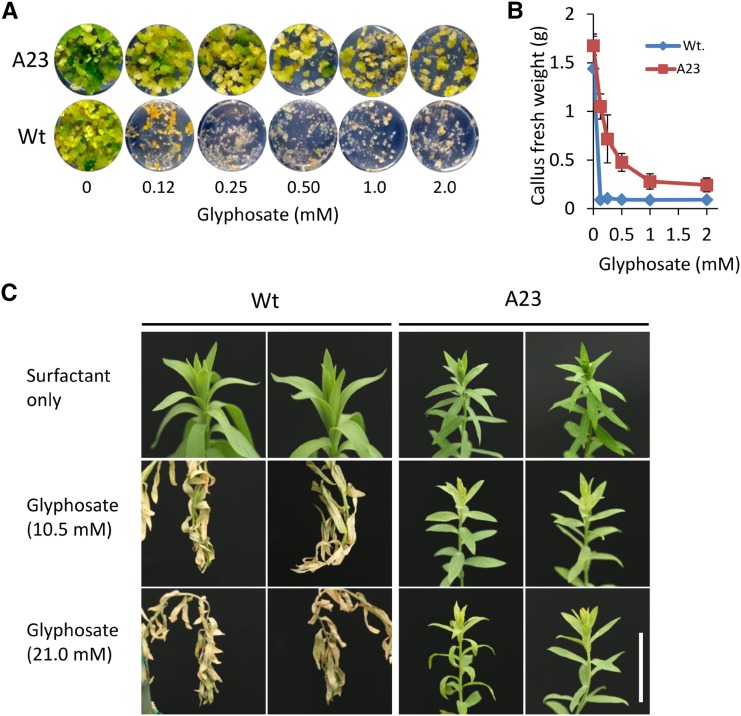
To determine the glyphosate tolerance afforded by the T178I and P182A mutations, we challenged callus line A23, a line that was identified as being heterozygous for the T178I and P182A edits in EPSPS gene 2, as well as the whole C0 plants regenerated from this callus line with glyphosate. A23 callus and control wild-type callus were plated on solid regeneration medium containing a range of glyphosate concentrations. After 21 d, the fresh weight of calli with T178I and P182A edits was significantly higher (P < 0.01) than that of wild-type calli at all glyphosate concentrations tested (Fig. 9, A and B). For regenerated whole plants, both wild-type and EPSPS edited plants were maintained in soil under greenhouse conditions and then sprayed with either 10.5 or 21 mm glyphosate. Six days post treatment, wild-type plants exhibited a wilted and necrotic phenotype typical of glyphosate toxicity for both application rates, whereas A23 plants with the edited EPSPS gene exhibited minimal phenotypic change (Fig. 9C). This result is notable as it implies that a single T178I and P182A edited EPSPS gene provides a level of tolerance much greater than that observed in the control plants.
Figure 9.
Targeted EPSPS edits provide herbicide tolerance to flax calli and regenerated plants. A, Flax wild-type (Wt) calli and calli derived from event A23, which contains T178I and P182A edits in EPSPS gene 2, were cultured in six-well dishes on medium containing a range of glyphosate concentrations. Images were captured 14 d after the initiation of treatment. Each image corresponds to one well. B, Mean fresh weight per well of wild-type and A23 calli treated with glyphosate after 21 d. Data represent means ± se (n = 3). C, Greenhouse-hardened wild-type and A23 whole plants in soil were treated with 10.5 or 21 mm glyphosate or surfactant only by spray application. Images were captured 6 d after glyphosate application. This experiment was repeated multiple times with similar results. Bar = 2 cm.
Taken together, these data demonstrate that, in flax, ssODNs combined with CRISPR/Cas9 can result in precise EPSPS edits at sufficient frequency to be detected by molecular screening without the need for selective culture conditions and that these edits are properly transmitted to subsequent generations.
DISCUSSION
Here we report, in two different plant systems, that precise scarless edits mediated by reagents that make DSBs can be enhanced significantly when combined with ssODNs. Delivery of ssODNs to Arabidopsis protoplasts pretreated with the nonspecific DSB reagent phleomycin improved the BFP-to-GFP editing frequency at doses higher than what has been reported previously for such antibiotics (Suzuki et al., 2003). We postulate that this difference in effective dose may be related to the membrane permeability of phleomycin in Arabidopsis protoplasts (Sidik and Smerdon, 1990). While effective at improving genome editing, it should be noted that, because of its nondiscriminant nature, there is potential for phleomycin to cause random DSBs remote from the target site that will be repaired imprecisely by NHEJ. These mutations, however, are analogous to those generated using a chemical mutagenesis approach typical of traditional plant breeding programs, and these mutations could, in most cases, be eliminated through outcrossing. As such, the use of ssODNs and phleomycin as a method for generating precise genome edits is notable and implies that the generation of DSBs is an important factor for improving genome-editing efficiencies in plants.
A more significant increase in the frequency of precise scarless genome edits was observed when ssODNs were used with the engineered nucleases TALENs or CRISPR/Cas9. We suspect that this increase is related to the generation of a target-specific DSB in close proximity to the intended edit site (Yang et al., 2013). In Arabidopsis protoplasts, CRISPR/Cas9 activity, as measured by indel mutations, was on average higher than in equivalent experiments with TALENs (Fig. 3C). A similar result was reported by Liang et al. (2014) when targeting the same locus in maize with both TALEN and CRISPR/Cas9. However, we cannot rule out other factors, such as transcriptional efficiency, engineered nuclease architecture, differences in target region, or methylation status, among others, that may contribute to this difference in targeting efficiency (Bortesi and Fischer, 2015). In our work, the frequency of precise genome edits was higher in CRISPR/Cas9-treated than in TALEN-treated Arabidopsis protoplasts when ssODNs were added. We interpret this result to be a function of the higher nuclease activity exhibited by CRISPR/Cas9 at the BFP target site. Notably, regardless of the engineered nuclease type, we consistently found that increased ssODN length had a positive effect on editing. A similar effect was observed in editing experiments without supplemental nuclease, suggesting that this enhancement is independent of DSBs near the target site. One would assume that this affect could be related to the higher level of homology to the target region that longer ssODNs possess, but it may be simply related to an increased in vivo half-life. More rigorous examination will be required to fully understand how ssODN length influences genome editing.
While a combinatorial approach to genome editing using engineered nucleases and ssODNs has been reported in fish and mammals (Ding et al., 2013; Hwang et al., 2013; Wefers et al., 2013; Yang et al., 2013; Strouse et al., 2014), there are few published data for plants. Shan et al. (2015), using a partial transgenic approach, used CRISPR/Cas9 and ssODNs in rice protoplasts to introduce two restriction enzyme sites in the OsPDS gene. However, the authors did not report the regeneration of whole plants from the successfully edited protoplasts. Employing TALENs and a chimeric RNA/DNA oligonucleotide (chimeraplast) to target the OsEPSPS gene, Wang et al. (2015) reported detecting one out of 25 transgenic lines with their intended gene edit. However, this line, in addition to the single base edit, also contained an imprecise NHEJ event. In maize, Svitashev et al. (2015) bombarded immature embryos with ssODNs and CRISPR/Cas9 targeting the ALS gene, a target for sulfonylurea herbicides. Using a double selection strategy, the authors first selected the transgenic lines that had integrated CRISPR/Cas9 reagents using a phosphinothricin acetyltransferase selectable marker that confers resistance to bialophos and then selected for the targeted events with the herbicide chlorsulfuron. Regenerated plants from successfully edited ALS lines were shown subsequently to be tolerant to herbicide.
Employing CRISPR/Cas9 in combination with ssODNs, we efficiently generated nontransgenic plants with precise scarless genome edits in each of the two flax EPSPS genes. Because we obtained a high edit frequency, we were able to regenerate whole fertile plants from edited protoplasts without the use of a selection agent. This is notable as it allows for the efficient targeting of nonselectable traits or non-protein-coding DNA such as promoters and noncoding RNA. To our knowledge, this is the first report describing the regeneration of whole plants that contain the intended precise genome edits from individual protoplasts without using selection. Our successful introduction of a glyphosate tolerance trait into flax through the use of ODM underscores the potential of developing new traits in agriculturally important crop plants in a nontransgenic manner.
MATERIALS AND METHODS
Construction of Engineered Nucleases
The design and construction of the TALEN expression construct BT-1 was based on previously described rules (Cermak et al., 2011). The target sequence was selected based on the gene-editing site and the repeat variable residue following the rules that NG, HD, NI, and NN recognize T, C, A, and G, respectively. The assembly of the TAL effector domain linked to the heterodimeric FokI domains was completed through a commercial service (GeneArt; Life Technologies). TALEN monomers were cloned between the MAS promoter and the pea (Pisum sativum) ribulose bisphosphate carboxylase (rbcE9) terminator and expressed as a coupled unit. For construction of the transient CRISPR-Cas9 expression plasmids BC-1 and EC-2, a higher plant codon-optimized SpCas9 gene containing an SV40 NLS at both the N and C termini and a 3× FLAG tag on the N terminus was synthesized as a series of GeneArt Strings (Life Technology), assembled, and then subsequently cloned downstream of the MAS promoter and upstream of the rbcsE9 terminator by the method of Gibson et al. (2009). Next, an sgRNA cassette consisting of a chimeric genomic RNA, whose expression is driven by the Arabidopsis (Arabidopsis thaliana) U6 promoter, was synthesized as GeneArt Strings and then shuttled into the Cas9-containing construct using the method of Gibson et al. (2009), forming pBCRISPR. To specify the chimeric sgRNA for the respective target sequence, pairs of DNA oligonucleotides encoding the protospacers for BC-1 and EC-2 (Supplemental Table S4) were annealed to generate short double-stranded fragments with 4-bp overhangs. The fragments were then ligated into BbsI-digested pBCRISPR to yield CRISPR/Cas9 constructs BC-1 and EC-2.
Oligonucleotides
All ssODNs used in this study were synthesized by Trilink Biotechnologies. All amplification primers were synthesized by IDT.
Cell Culture and Protoplast Isolation
Arabidopsis
Surface-sterilized Arabidopsis seeds were germinated under sterile conditions. Roots from 2- to 3-week-old seedlings were collected and maintained in one-half-strength Murashige and Skoog (MS) liquid medium (Murashige and Skoog, 1962) under low light at 25°C. Cultures were transferred to and maintained in MSAR1.1 (MSAR1 as described in Mathur et al., 1995) without 6-(γ,γ-dimethylallylamino)purine riboside 3 weeks prior to protoplast isolation to induce root meristematic tissue. Root meristematic tissue was incubated in an enzyme solution containing 1.25% (w/v) Cellulase RS and 0.25% (w/v) Macerozyme R-10 with gentle shaking. The released protoplasts were collected and purified by density centrifugation. Protoplasts were resuspended in TM solution (14.8 mm MgCl2·6H2O, 5 mm MES, and 572 mm mannitol, pH 5.8).
Flax
Flax (Linum usitatissimum) protoplasts were isolated from 3-week-old seedlings germinated in vitro. After digestion of shoot tips in a solution containing 0.66% (w/v) Cellulase YC and 0.16% (w/v) Macerozyme R-10, the resulting protoplasts were purified by density centrifugation and used for transfection on the day following purification.
Protoplast Transfection
Arabidopsis protoplasts (2.5 × 105) were transfected with either 25 pmol of ssODN alone, 25 pmol of ssODN plus 5 µg of CRISPR-Cas9 plasmid (BC-1), 25 pmol of ssODN plus 5 µg of TALEN plasmid (BT-1), or mock treated using polyethylene glycol (PEG)-mediated delivery. Transfection occurred on ice for 10 min. Transfected protoplasts were cultured in MSAP (MSAR1.1 containing 0.4 m mannitol) in low light at 25°C. For experiments with phleomycin (InvivoGen), protoplasts were kept in TM adjusted to pH 7 for 90 min on ice before transfection. For flax, after 18 h of culture, 1 × 106 protoplasts were transfected with 200 pmol of ssODN and 20 µg of CRISPR-Cas9 plasmid (EC-2) using PEG-mediated delivery. Treated protoplasts were incubated in the dark at 25°C for up to 24 h, embedded in alginate beads (Roger et al., 1996), and cultured in basal V-KM liquid medium (Binding and Nehls, 1977) supplemented with 91 nm thidiazuron and 11 nm NAA. Edits to the EPSPS gene were assessed by NGS in genomic DNA extracted from pools of approximately 10,000 microcolonies obtained from protoplasts 3 and 7 weeks after transfection. Microcalli were then released from the alginate and transferred to solidified medium (MS salts, Morel and Wetmore vitamins [Morel and Wetmore, 1951], 3% [w/v] Suc, 91 nm thidiazuron, 11 nm NAA, pH 5.8, and 0.3% phytagel) for shoot regeneration at a density of 0.5 mL settled cell volume per plate. After about 3 weeks, individual calli (approximately 0.5 cm diameter) were split in two. One half was used for molecular screening, and the other half was kept on a 24-well plate. Shoots began to develop from calli after approximately 4 to 6 weeks. Elongated shoots were micropropagated and rooted in MS medium, and rooted plants were transferred to soil and hardened in a growth chamber for 2 to 4 weeks until the plants were well established.
Detection of Arabidopsis BFP-to-GFP Edits
Protoplasts were analyzed using the Attune Acoustic Focusing Cytometer (Applied Biosystems) with excitation and emission settings appropriate for GFP. Background fluorescence was based on PEG-treated protoplasts without DNA delivery. Data were normalized for transfection efficiency using a fluorescent protein reporter construct.
Gene Edit and Indel Sequencing
Genomic DNA was extracted from treated protoplasts using the NucleoSpin Plant II kit according to the manufacturer’s recommendations (Machery-Nagel). Amplicons were generated with primers flanking the BT-1 TALEN and BC-1 and EC-2 CRISPR/Cas9 target regions (Supplemental Fig. S1; Supplemental Table S3) using Phusion polymerase (New England Biolabs) and 100 ng of genomic DNA. The amplicons were purified and concentrated using Qiaquick MinElute columns (Qiagen) and then deep sequenced using a 2 × 250-bp MiSeq run (Illumina). For data analysis, FASTQ files for read 1 and read 2 were imported into the CLC Genomics Workbench 7.0.4 (CLCBio). Paired reads were merged into a single sequence if their sequences overlapped. A sequence for an amplicon was identified if it or its reverse and complemented sequence contained both forward and reverse primer sequences. The occurrence of a unique sequence in a sample was recorded as its abundance. The percentage of indels or targeted edits was calculated by dividing the number of sequences with the edit or indel by the total number of sequences and then multiplying by 100. For flax samples, genomic DNA from microcolony or callus samples was extracted using the EvoPURE plant DNA kit (Aline Biosciences) and screened by deep sequencing and allele-specific quantitative PCR (Morlan et al., 2009). Positive-scoring PCR fragments were then TOPO-TA cloned into the pCR2.1 vector (Invitrogen) according to the manufacturer’s protocol. Typically, cloned PCR fragments from 10 to 15 transformants were then TempliPhi sequenced (GE Healthcare Life Sciences) to confirm the DNA sequence for each EPSPS allele from a single isolated callus. A similar PCR cloning and sequencing procedure was used for DNA sequence confirmation in leaf samples of regenerated shoots.
Off-Target Analysis
Potential off-target loci for BC-1 in the Arabidopsis genome were determined using Cas-OFFinder (Bae et al., 2014). Potential off-target loci for EC-2 were determined by BLAST searches of the flax genome database in Phytozyme 10.2 (http://phytozome.jgi.doe.gov). Off-target sites based on sequence identities to the protospacer were screened for mutations by deep sequencing. Genomic DNA was extracted from Arabidopsis protoplasts or flax regenerated plants and amplicons generated with Phusion polymerase using primers that flank the potential off-target site (Supplemental Table S3). The amplicons were subjected to deep sequencing using a 2 × 250-bp MiSeq run. Mutations near the expected cleavage site were considered off-target events. The percentage of indels was calculated by dividing the number of sequences with the indel by the total number of sequences and then multiplying by 100.
Herbicide Tolerance Tests
The glyphosate tolerance of calli and regenerated plants was assessed in vitro and in the greenhouse, respectively. Individual calli were cloned by cutting and culturing smaller pieces in fresh regeneration medium to increase callus mass. Calli derived from wild-type leaf protoplasts were used as negative controls. Calli derived from subcultures of individual callus lines were then pooled and broken up into 0.5- to 1-mm pieces by blending in liquid MS medium (4.33 g L−1 MS salts, 3% [w/v] Suc, and 0.1% [w/v] Morel and Wetmore vitamins), and 0.25 mL of settled callus pieces was inoculated on regeneration medium containing 0, 0.125, 0.25, 0.5, 1, or 2 mm glyphosate in six-well dishes. Treatments were performed in triplicate (three wells per treatment), and the experiments were repeated three times. Prior to spray tests, regenerated plants were subjected to a hardening period in a growth chamber under a 16-h photoperiod with day and night temperatures of 21°C and 18°C, respectively. Hardened plants regenerated from callus were transferred to the greenhouse for glyphosate treatment. Wild-type control and EPSPS edited plants were sprayed with 10.5 or 21 mm glyphosate (Roundup Pro; Monsanto). Treatment rates were normalized to a spray volume of 75.7 liters per acre to replicate field conditions. A mock treatment of surfactant only was included as a control. Plants were evaluated and photographed 6 d after the glyphosate treatment to determine herbicide tolerance.
Statistical Analysis
Statistical significance was determined using a Student’s t test with two-tailed distribution. P < 0.05 was considered significant. Data are shown as means ± se. The fresh weight of calli grown in vitro under different glyphosate concentrations was analyzed using ANOVA and Tukey’s test for mean separation.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Diagram showing the methodology for amplicon deep sequencing analysis.
Supplemental Table S1. Sequences of ssODNs used in this study.
Supplemental Table S2. Sequences of TALE binding domains.
Supplemental Table S3. Primers used in this study.
Supplemental Table S4. Protospacer sequences of sgRNAs used in this study.
Supplemental Table S5. Analysis of BC-1 off-targets.
Supplemental Table S6. Analysis of off-targets for flax plant A23.
Supplemental Text S1. Sequences and locus names for flax EPSPS gene 1 and gene 2.
Supplementary Material
Acknowledgments
We thank all of the members of Cibus for their many contributions to this work (Olga Batalov, Corey Cournoyer, Daniel Galindo, Miranda Marks, Amber Perry, Douglas Potter, Reema Sāleh, Derek Sanford, and Andrew Walker) as well as Joseph Ecker (Salk Institute for Biological Studies) and Stephen Howell (Iowa State University) for critical review of the article.
Glossary
- ODM
oligonucleotide-directed mutagenesis
- DSB
double strand break
- CRISPR/Cas9
clustered, regularly interspaced, short palindromic repeat
- NHEJ
nonhomologous end joining
- indel
deletions and insertions
- ssODN
single-stranded oligonucleotide
- sgRNA
single guide RNA
- PAM
protospacer adjacent motif
- NLS
nuclear localization signal
- MS
Murashige and Skoog
- PEG
polyethylene glycol
Footnotes
Articles can be viewed without a subscription.
References
- Aarts M, Dekker M, de Vries S, van der Wal A, te Riele H (2006) Generation of a mouse mutant by oligonucleotide-mediated gene modification in ES cells. Nucleic Acids Res 34: e147–e149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrieu-Soler C, Casas M, Faussat AM, Gandolphe C, Doat M, Tempé D, Giovannangeli C, Behar-Cohen F, Concordet JP (2005) Stable transmission of targeted gene modification using single-stranded oligonucleotides with flanking LNAs. Nucleic Acids Res 33: 3733–3742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae S, Park J, Kim JS (2014) Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 30: 1473–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beetham PR, Kipp PB, Sawycky XL, Arntzen CJ, May GD (1999) A tool for functional plant genomics: chimeric RNA/DNA oligonucleotides cause in vivo gene-specific mutations. Proc Natl Acad Sci USA 96: 8774–8778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialk P, Rivera-Torres N, Strouse B, Kmiec EB (2015) Regulation of gene editing activity directed by single-stranded oligonucleotides and CRISPR/Cas9 systems. PLoS ONE 10: e0129308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binding H, Nehls R (1977) Regeneration of isolated protoplasts to plants in Solanum dulcamara L. Z Pflanzenphysiol 85: 279–280 [Google Scholar]
- Bortesi L, Fischer R (2015) The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol Adv 33: 41–52 [DOI] [PubMed] [Google Scholar]
- Carlson DF, Tan W, Lillico SG, Stverakova D, Proudfoot C, Christian M, Voytas DF, Long CR, Whitelaw CB, Fahrenkrug SC (2012) Efficient TALEN-mediated gene knockout in livestock. Proc Natl Acad Sci USA 109: 17382–17387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF (2011) Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res 39: e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Piédoue G, Andrieu-Soler C, Concordet JP, Maurisse R, Sun JS, Lopez B, Kuzniak I, Leboulch P, Feugeas JP (2007) Targeted gene correction with 5′ acridine-oligonucleotide conjugates. Oligonucleotides 17: 258–263 [DOI] [PubMed] [Google Scholar]
- Ding Q, Lee YK, Schaefer EA, Peters DT, Veres A, Kim K, Kuperwasser N, Motola DL, Meissner TB, Hendriks WT, et al. (2013) A TALEN genome-editing system for generating human stem cell-based disease models. Cell Stem Cell 12: 238–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA III, Smith HO (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6: 343–345 [DOI] [PubMed] [Google Scholar]
- Gocal FW, Schöpke C, Beetham PR (2015) Oligo-mediated targeted gene editing. In Zhang F, Puchta H, Thomson JG, eds, Advances in New Technology for Targeted Modification of Plant Genomes. Springer, New York, pp 73–90 [Google Scholar]
- Gocal G, Knuth M, Beetham P, inventors. January 10, 2007. Generic EPSPS mutants. U.S. Patent Application No. US8268622
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, et al. (2013) DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 31: 827–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK (2013) Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol 31: 227–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z, Zhang K, Chen K, Gao C (2014) Targeted mutagenesis in Zea mays using TALENs and the CRISPR/Cas system. J Genet Genomics 41: 63–68 [DOI] [PubMed] [Google Scholar]
- Lor VS, Starker CG, Voytas DF, Weiss D, Olszewski NE (2014) Targeted mutagenesis of the tomato PROCERA gene using transcription activator-like effector nucleases. Plant Physiol 166: 1288–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur J, Koncz C, Szabados L (1995) A simple method for isolation, liquid culture, transformation and regeneration of Arabidopsis thaliana protoplasts. Plant Cell Rep 14: 221–226 [DOI] [PubMed] [Google Scholar]
- Menke M, Chen I, Angelis KJ, Schubert I (2001) DNA damage and repair in Arabidopsis thaliana as measured by the comet assay after treatment with different classes of genotoxins. Mutat Res 493: 87–93 [DOI] [PubMed] [Google Scholar]
- Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, et al. (2011) A TALE nuclease architecture for efficient genome editing. Nat Biotechnol 29: 143–148 [DOI] [PubMed] [Google Scholar]
- Moerschell RP, Tsunasawa S, Sherman F (1988) Transformation of yeast with synthetic oligonucleotides. Proc Natl Acad Sci USA 85: 524–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel G, Wetmore R (1951) Fern callus tissue culture. Am J Bot 38: 141–143 [Google Scholar]
- Morlan J, Baker J, Sinicropi D (2009) Mutation detection by real-time PCR: a simple, robust and highly selective method. PLoS ONE 4: e4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog FA (1962) A revised medium for rapid growth and bio-assays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Olsen PA, Randol M, Krauss S (2005) Implications of cell cycle progression on functional sequence correction by short single-stranded DNA oligonucleotides. Gene Ther 12: 546–551 [DOI] [PubMed] [Google Scholar]
- Radecke S, Radecke F, Peter I, Schwarz K (2006) Physical incorporation of a single-stranded oligodeoxynucleotide during targeted repair of a human chromosomal locus. J Gene Med 8: 217–228 [DOI] [PubMed] [Google Scholar]
- Rios X, Briggs AW, Christodoulou D, Gorham JM, Seidman JG, Church GM (2012) Stable gene targeting in human cells using single-strand oligonucleotides with modified bases. PLoS ONE 7: e36697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roger D, David A, David H (1996) Immobilization of flax protoplasts in agarose and alginate beads: correlation between ionically bound cell-wall proteins and morphogenetic response. Plant Physiol 112: 1191–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer NJ, Mozoruk J, Miller RB, Warburg ZJ, Walker KA, Beetham PR, Schöpke CR, Gocal GF (2016) Oligonucleotide-directed mutagenesis for precision gene editing. Plant Biotechnol J 14: 496–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönbrunn E, Eschenburg S, Shuttleworth WA, Schloss JV, Amrhein N, Evans JN, Kabsch W (2001) Interaction of the herbicide glyphosate with its target enzyme 5-enolpyruvylshikimate 3-phosphate synthase in atomic detail. Proc Natl Acad Sci USA 98: 1376–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröpfer S, Knoll A, Trapp O, Puchta H (2014) DNA repair and recombination in plants. In Howell SH, ed, Molecular Biology. Springer, New York, pp 51–93 [Google Scholar]
- Shan Q, Zhang Y, Chen K, Zhang K, Gao C (2015) Creation of fragrant rice by targeted knockout of the OsBADH2 gene using TALEN technology. Plant Biotechnol J 13: 791–800 [DOI] [PubMed] [Google Scholar]
- Sidik K, Smerdon MJ (1990) Bleomycin-induced DNA damage and repair in human cells permeabilized with lysophosphatidylcholine. Cancer Res 50: 1613–1619 [PubMed] [Google Scholar]
- Strouse B, Bialk P, Niamat RA, Rivera-Torres N, Kmiec EB (2014) Combinatorial gene editing in mammalian cells using ssODNs and TALENs. Sci Rep 4: 3791–3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Murai A, Muramatsu T (2003) Low-dose bleomycin induces targeted gene repair frequency in cultured melan-c cells using chimeric RNA/DNA oligonucleotide transfection. Int J Mol Med 12: 109–114 [PubMed] [Google Scholar]
- Svitashev S, Young JK, Schwartz C, Gao H, Falco SC, Cigan AM (2015) Targeted mutagenesis, precise gene editing, and site-specific gene insertion in maize using Cas9 and guide RNA. Plant Physiol 169: 931–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voytas DF. (2013) Plant genome engineering with sequence-specific nucleases. Annu Rev Plant Biol 64: 327–350 [DOI] [PubMed] [Google Scholar]
- Wang M, Liu Y, Zhang C, Liu J, Liu X, Wang L, Wang W, Chen H, Wei C, Ye X, et al. (2015) Gene editing by co-transformation of TALEN and chimeric RNA/DNA oligonucleotides on the rice OsEPSPS gene and the inheritance of mutations. PLoS ONE 10: e0122755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wefers B, Meyer M, Ortiz O, Hrabé de Angelis M, Hansen J, Wurst W, Kühn R (2013) Direct production of mouse disease models by embryo microinjection of TALENs and oligodeoxynucleotides. Proc Natl Acad Sci USA 110: 3782–3787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Guell M, Byrne S, Yang JL, De Los Angeles A, Mali P, Aach J, Kim-Kiselak C, Briggs AW, Rios X, et al. (2013) Optimization of scarless human stem cell genome editing. Nucleic Acids Res 41: 9049–9061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon K, Cole-Strauss A, Kmiec EB (1996) Targeted gene correction of episomal DNA in mammalian cells mediated by a chimeric RNA.DNA oligonucleotide. Proc Natl Acad Sci USA 93: 2071–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Gou F, Zhang J, Liu W, Li Q, Mao Y, Botella JR, Zhu JK (2015) TALEN-mediated targeted mutagenesis produces a large variety of heritable mutations in rice. Plant Biotechnol J 14: 186–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu T, Peterson DJ, Tagliani L, St Clair G, Baszczynski CL, Bowen B (1999) Targeted manipulation of maize genes in vivo using chimeric RNA/DNA oligonucleotides. Proc Natl Acad Sci USA 96: 8768–8773 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.