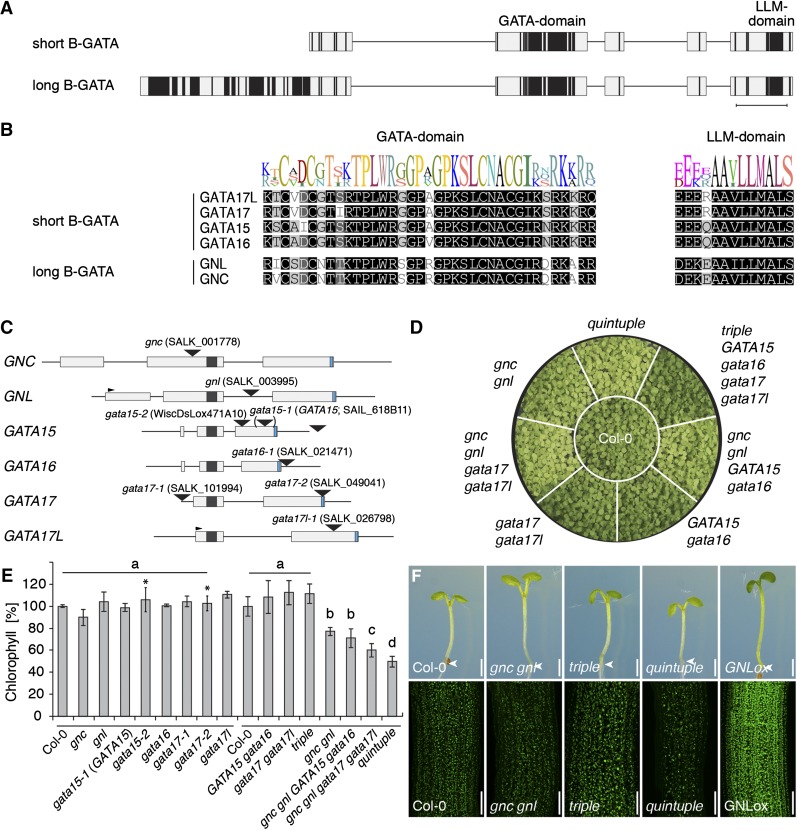
Figure 1.
LLM-domain B-GATAs redundantly control greening in Arabidopsis. A, Domain architecture of short and long LLM-domain B-GATAs based on the alignment of family members from Arabidopsis. Protein regions with restricted sequence conservation or gaps in the alignment are depicted as lines; regions with low (≤50%) sequence similarity, gray boxes; regions with high (≥80%) sequence similarity, black boxes. Scale bar = 30 amino acids in a conserved part of the protein. B, MUSCLE alignment with sequence logo of the B-GATA domain and the LLM-domain of the Arabidopsis LLM-domain B-GATAs. C, Schematic representation of the gene models of the six Arabidopsis LLM-domain B-GATA genes with exons (boxes) and introns (lines). The 5′-UTR (untranslated region) of GNL and the 3′-UTR of GATA16 were previously unknown and derived from RNA sequencing data available in the laboratory. The sequences coding for the GATA DNA-binding domain and the LLM-domain are represented as black and blue boxes, respectively. The T-DNA insertion positions are shown. The position in brackets of the GATA15 insertion refers to the original misannotation of this insertion (www.signal.salk.edu). D, Photograph of 14-d-old plants with the genotypes as specified in comparison to the Columbia-0 (Col-0) wild-type control. E, Result of the quantification of chlorophyll A and B from different gata single and complex mutants. The experiment was performed in two rounds, as indicated by the lines above the genotypes, to reduce the complexity of the experiment; the Col-0 wild-type reference was included in both rounds. F, Representative photographs of 7-d-old seedlings (top panel) and confocal images (bottom panel) of these seedlings to visualize differential chloroplast accumulation in these mutants.