Micro-RNA miR444 and its MADS-box targets directly alter RNA-dependent RNA polymerase transcription in antiviral defense.
Abstract
Plant RNA-DEPENDENT RNA POLYMERASE1 (RDR1) is a key component of the antiviral RNA-silencing pathway, contributing to the biogenesis of virus-derived small interfering RNAs. This enzyme also is responsible for producing virus-activated endogenous small interfering RNAs to stimulate the broad-spectrum antiviral activity through silencing host genes. The expression of RDR1 orthologs in various plants is usually induced by virus infection. However, the molecular mechanisms of activation of RDR1 expression in response to virus infection remain unknown. Here, we show that a monocot-specific microRNA, miR444, is a key factor in relaying the antiviral signaling from virus infection to OsRDR1 expression. The expression of miR444 is enhanced by infection with Rice stripe virus (RSV), and overexpression of miR444 improves rice (Oryza sativa) resistance against RSV infection accompanied by the up-regulation of OsRDR1 expression. We further show that three miR444 targets, the MIKCC-type MADS box proteins OsMADS23, OsMADS27a, and OsMADS57, form homodimers and heterodimers between them to repress the expression of OsRDR1 by directly binding to the CArG motifs of its promoter. Consequently, an increased level of miR444 diminishes the repressive roles of OsMADS23, OsMADS27a, and OsMADS57 on OsRDR1 transcription, thus activating the OsRDR1-dependent antiviral RNA-silencing pathway. We also show that overexpression of miR444-resistant OsMADS57 reduced OsRDR1 expression and rice resistance against RSV infection, and knockout of OsRDR1 reduced rice resistance against RSV infection. In conclusion, our results reveal a molecular cascade in the rice antiviral pathway in which miR444 and its MADS box targets directly control OsRDR1 transcription.
RNA silencing mediated by regulatory small RNAs (microRNAs [miRNAs] and small interfering RNAs [siRNAs]) negatively regulates gene expression at the posttranscriptional level or at the transcriptional level in eukaryotic organisms. Besides small RNAs, plant RNA-silencing pathways incorporate several kinds of core protein components, such as DICER-LIKE (DCL) RNase III endonucleases, which process long double-stranded RNA (dsRNA) into small RNA duplexes; ARGONAUTEs (AGOs), the major effector of the RNA-induced silencing complexes, which bind to small RNAs for silencing target RNAs; and RNA-dependent RNA polymerases (RDRs), which are required for copying single-stranded RNAs into dsRNAs for downstream processing by DCLs. Multiple DCLs, AGOs, and RDRs have evolved in plants and thus form an array of RNA-silencing pathways (Axtell, 2013; Martínez de Alba et al., 2013; Bologna and Voinnet, 2014). Among them, the antiviral RNA-silencing pathway is the earliest described and most extensively studied. It is well known that the antiviral silencing pathway directly targets viral RNAs. Briefly, as in Arabidopsis (Arabidopsis thaliana), the stem-loop structures and dsRNA replication intermediates of viral RNAs are recognized and cleaved by DCLs (DCL4 and DCL2) to produce primary virus-derived small interfering RNAs (vsiRNAs). Furthermore, abundant secondary vsiRNAs are produced from the dsRNAs amplified by host-encoded RDRs (RDR1 or RDR6). Both primary and secondary vsiRNAs are loaded into AGO proteins (AGO1 and AGO2) to direct the degradation of the viral RNAs. The amplified vsiRNAs are believed also to trigger the systemic silencing of the viral RNAs in distant tissues (Ding and Voinnet, 2007; Ding, 2010; Pumplin and Voinnet, 2013).
On the other hand, recent studies have shown that antiviral RNA silencing also is attributed to alterations in the expression of host genes, including those directly involved in the antiviral RNA-silencing pathway. For example, DCL1, a target of microRNA162 (miR162), negatively regulated the expression of DCL4 and DCL3, two important DCLs for slicing viral RNAs and producing vsiRNAs (Qu et al., 2008; Azevedo et al., 2010). Also, the activation of AGO1 expression has been observed in Arabidopsis infected by different viruses (Zhang et al., 2006; Azevedo et al., 2010; Várallyay et al., 2010), and AGO2 expression increased in Turnip crinkle virus- and Cucumber mosaic virus-infected plants (Harvey et al., 2011). The induction of these genes would certainly enhance the function of antiviral RNA-silencing pathways.
In addition, silencing of other defense-related host genes contributes to the antiviral response. For example, virus infection reduced the accumulation of miR482, a 22-nucleotide miRNA targeting the nucleotide-binding site-leucine-rich repeat (NBS-LRR) class R genes, and consequently increased the expression of two miR482-targeted NBS-LRR mRNAs in infected plants (Shivaprasad et al., 2012). And down-regulation of an R gene due to increased expression of miR6019, another 22-nucleotide miRNA, resulted in the attenuated R gene-mediated resistance to Tobacco mosaic virus in Nicotiana benthamiana (Li et al., 2012). Similarly, Turnip mosaic virus infection in Brassica spp. induced the production of miR1885, which targeted the TIR-NBS-LRR class R genes (He et al., 2008). In addition to miRNAs, siRNA-mediated RNA silencing also is involved in host defense against virus infection. For instance, a miniature inverted repeat transposable element inserted in the third intron of the tobacco mosaic virus resistance gene N generated 24-nucleotide siRNAs to regulate the expression of the N gene via an RNA-directed DNA methylation-mediated RNA-silencing mechanism (Kuang et al., 2009).
RDR1 orthologs have been reported to be virus or salicylic acid inducible in different plants, including Arabidopsis, Nicotiana spp., Medicago truncatula, maize (Zea mays), and rice (Oryza sativa; Yang et al., 2004; Alamillo et al., 2006; He et al., 2010; Satoh et al., 2010; Du et al., 2011), and to provide basal resistance to several viruses by participating in the biogenesis of virus-derived secondary siRNAs (Diaz-Pendon et al., 2007; Qi et al., 2009; Garcia-Ruiz et al., 2010; Wang et al., 2010). Recently, it was reported that RDR1 also is responsible for the production of a distinct class of virus-activated siRNAs, which direct widespread silencing of host genes to confer broad-spectrum antiviral activity in Arabidopsis (Cao et al., 2014), indicating that RDR1 plays a key role in antiviral resistance by silencing both the viral RNAs and the host immunity-related genes. Rice OsRDR1 might have similar roles in antiviral RNA silencing (Chen et al., 2010; Wang et al., 2014). However, the signaling pathway from virus infection to RDR1 expression is unclear.
miR444 is specific to monocots and plays roles in rice tillering and nitrate signaling (Guo et al., 2013; Yan et al., 2014). miR444 is a kind of natural antisense miRNA and targets four MIKCC-type MADS box homologous genes (OsMADS23, OsMADS27a, OsMADS27b, and OsMADS57) in rice (Sunkar et al., 2005; Lu et al., 2008; Wu et al., 2009; Li et al., 2010; Yan et al., 2014). Plant MADS box proteins regulate gene expression by binding to a highly conserved DNA motif known as the CArG box using the MADS box of the DNA-binding domain (de Folter and Angenent, 2006; Ito et al., 2008; Fujisawa et al., 2013). To bind the CArG motif, MADS box transcription factors need to form homodimeric or heterodimeric complexes (Riechmann et al., 1996). Here, we show that miR444 plays key roles in relaying the antiviral signal from virus infection to OsRDR1 expression in rice plants. Infection with Rice stripe virus (RSV) induces miR444 accumulation, and the activation of miR444 results in an increase in OsRDR1 expression, leading to rice resistance to RSV infection. We reveal a regulatory mechanism for the activation of OsRDR1 gene transcription in which the repressors formed by miR444 target proteins are released from the OsRDR1 promoter upon RSV infection. To our knowledge, such a derepression mechanism has not been described in the regulation of other RDR1 ortholog genes.
RESULTS
miR444 Accumulation Is Induced by Virus Infection in Rice
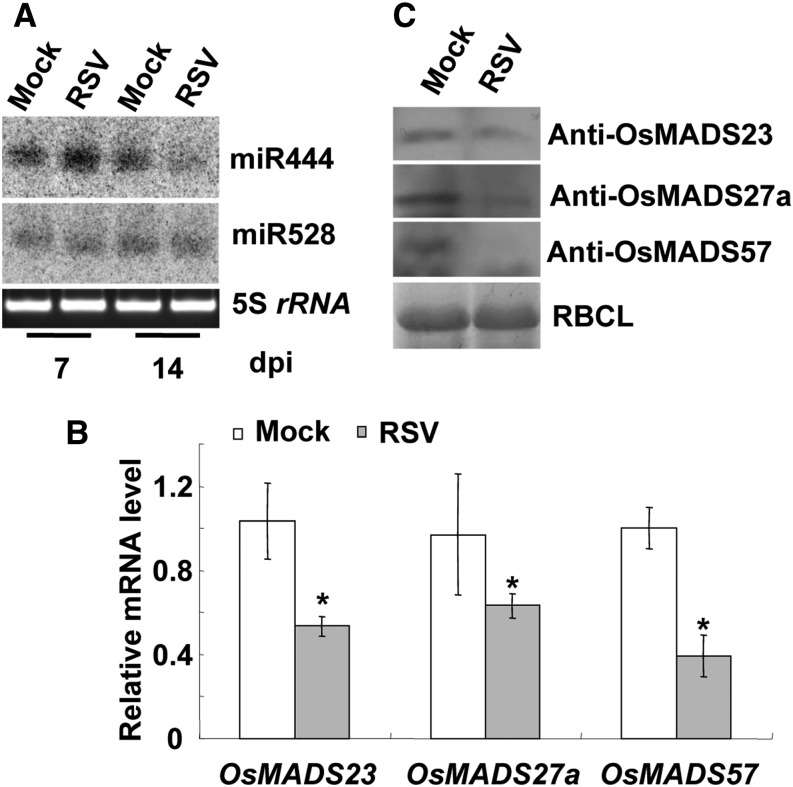
To study the potential function of miR444 in the interaction between rice and viral pathogens, we first analyzed the expression pattern of miR444 in RSV-challenged rice plants. To do this, RSV-susceptible cv Nipponbare plants were inoculated by RSV, then miR444 accumulation was estimated by small RNA gel blot at 7 and 14 d post inoculation (dpi). The results showed that miR444 accumulation was increased at 7 dpi (Fig. 1A). In contrast, miR528 accumulation remained unchanged between mock- and RSV-inoculated plants (Fig. 1A), suggesting that miR444 is especially induced in rice during RSV infection. We also determined the mRNA and protein accumulation of the miR444 targets in RSV-infected rice plants (7 dpi). These results showed that the mRNA and protein levels of OsMADS23, OsMADS27a, and OsMADS57 decreased to different extents (Fig. 1, B and C), consistent with the increased miR444 levels after RSV infection (Fig. 1A).
Figure 1.
miR444 accumulation was induced by virus infection. A, Analysis of the accumulation of miR444 and miR528 by RNA gel-blot hybridization in RSV- and mock-infected rice plants. Total RNA samples for hybridization were collected at 7 and 14 dpi. The 5S ribosomal RNA (rRNA) bands were visualized by ethidium bromide staining and served as a loading control. B, Quantitative reverse transcription (qRT)-PCR analysis of the expression of OsMADS23, OsMADS27a, and OsMADS57 in RSV-infected and mock control rice plants. After RT from the total RNA samples described in A (7 dpi), the relative mRNA levels were determined by qRT-PCR. Results are means ± se for three replicates. Asterisks indicate significant differences between infected and mock control rice plants. (Student’s t test analysis, P < 0.05). C, The accumulation of OsMADS23, OsMADS27a, and OsMADS57 proteins was analyzed by western blot at 7 dpi of RSV infection. The Rubisco large subunit (RBCL) bands were visualized by Coomassie Brilliant Blue and served as a loading control.
Overexpression of miR444 Increases Rice Resistance against Virus Infection
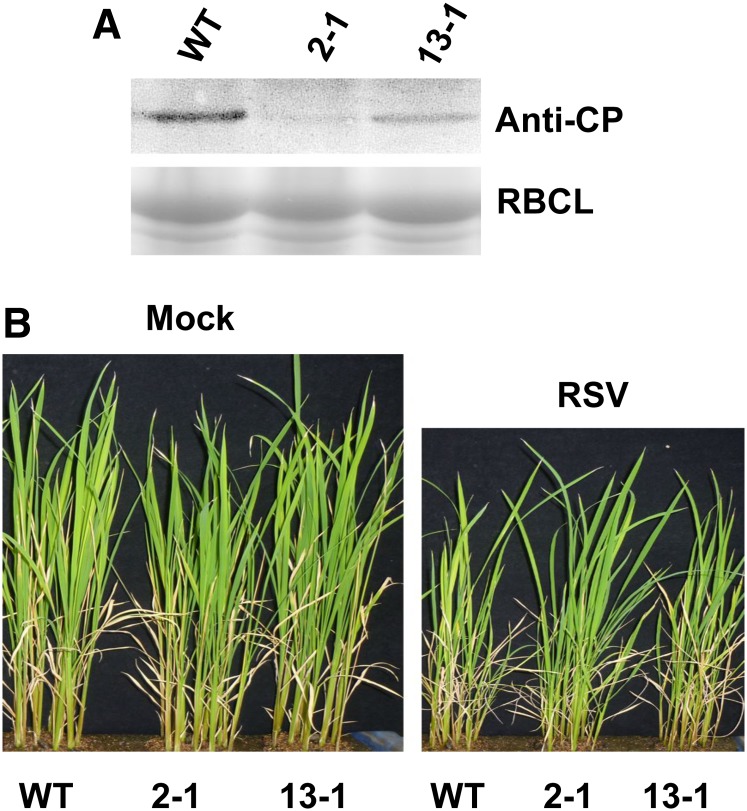
We then used two miR444-overexpressed rice lines (2-1 and 13-1) of cv Nipponbare (previously described in Yan et al., 2014) to evaluate the effect of miR444 up-regulation on the rice response to RSV infection. To do this, wild-type, 2-1, and 13-1 rice plants were grown in soil, and the seedlings were challenged with RSV. RSV NUCLEOCAPSID PROTEIN (CP) accumulation and plant growth were comparably analyzed. The results showed that miR444-overexpressed lines accumulated less RSV CP than wild-type rice plants at 14 dpi (Fig. 2A). At 1 month after infection, wild-type rice plants exhibited more severe chlorotic and dwarf symptoms in comparison with the miR444-overexpressed lines (Fig. 2B). Taken together, these observations indicate that the induction of miR444 expression has a positive role in rice resistance against RSV infection.
Figure 2.
Overexpression of miR444 improves rice resistance against RSV infection. A, Wild-type (WT) and miR444-overexpressed (2-1 and 13-1) rice plants were grown for 14 d and then inoculated by RSV. Protein samples were collected, and the accumulation of RSV CP was analyzed by western blot at 14 dpi of RSV infection. The Rubisco large subunit (RBCL) bands were visualized by Coomassie Brilliant Blue and served as a loading control. B, Growth performance of wild-type and miR444-overexpressed rice plants at 30 dpi of RSV infection.
miR444 Up-Regulates the Expression of OsRDR1 Constitutively and during Virus Infection
To further explore the regulatory mechanisms of miR444-mediated resistance against RSV infection, preliminary RNA sequencing (RNA-seq) was performed to analyze the global gene expression pattern in the two miR444-overexpressed rice lines (2-1 and 13-1). Interestingly, OsRDR1 (LOC_Os02g50330), a key gene in the antiviral RNA-silencing pathway, was up-regulated in both miR444-overexpressed rice lines (Supplemental Table S1). A further qRT-PCR analysis found that the level of the OsRDR1 mRNA was approximately 2.5- and 4-fold higher in miR444-overexpressed rice plants than in wild-type plants (Fig. 3A). It has been reported that RSV infection induced the expression of OsRDR1 in rice (Satoh et al., 2010; Du et al., 2011). As we have shown that RSV infection induced the accumulation of miR444 (Fig. 1A), it is reasonable to suggest that the increased expression of OsRDR1 in RSV-infected rice is mediated by miR444. Consistently, after RSV infection, the levels of the OsRDR1 mRNA are much higher in miR444-overexpressed rice plants than in wild-type rice plants (Fig. 3B). These results indicate that miR444 positively regulates the expression of OsRDR1 constitutively and more strongly during virus infection. Taken together, the above observations suggest that miR444-mediated virus resistance involved the activation of the OsRDR1-dependent antiviral RNA-silencing pathway.
Figure 3.
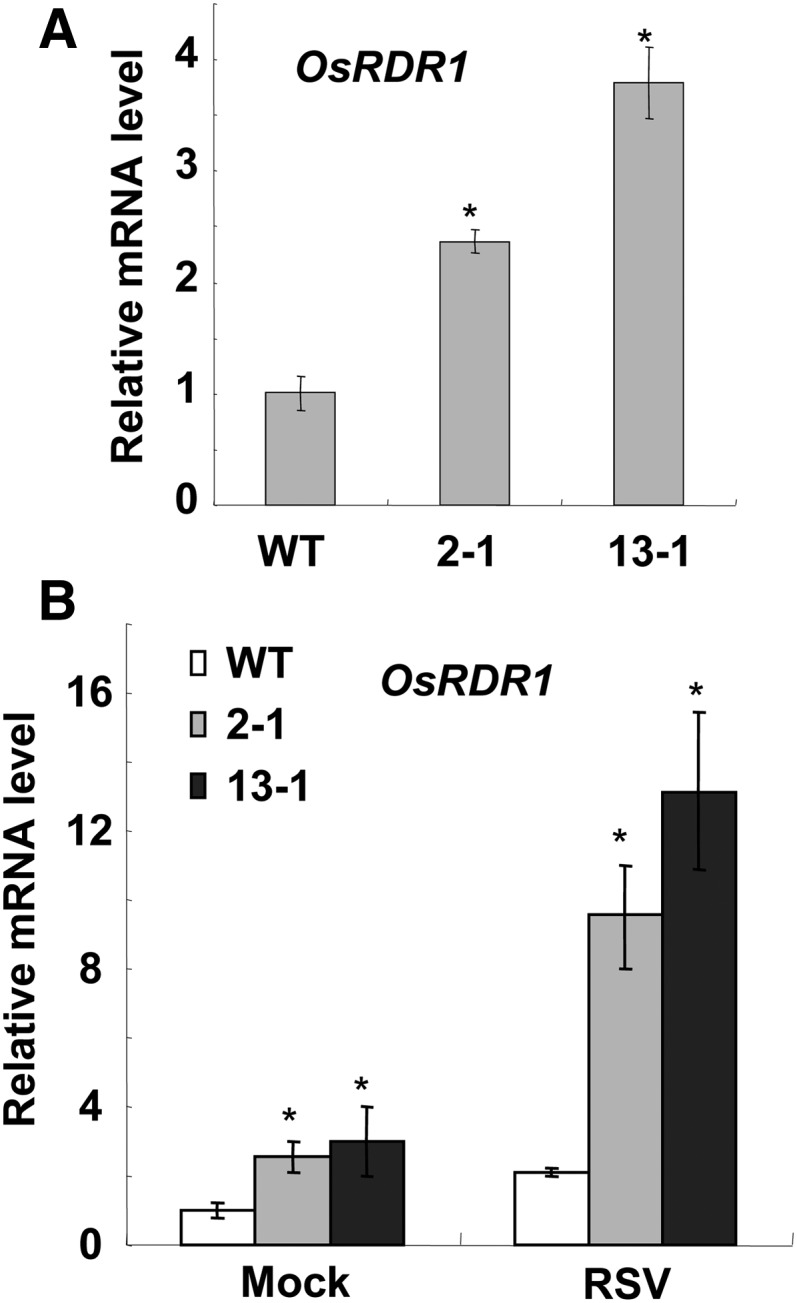
miR444 positively regulates the expression of OsRDR1 constitutively and upon virus infection. A, Overexpression of miR444 increased the expression of OsRDR1 constitutively. RNA samples were collected from wild-type (WT) and miR444-overexpressed rice plants (grown for 14 d). OsRDR1 mRNA levels relative to that of the wild-type rice plant were determined by qRT-PCR. B, Overexpression of miR444 markedly increased the expression of OsRDR1 during virus infection. Wild-type and miR444-overexpressed rice plants (grown for 14 d) were inoculated by RSV. At 14 dpi, RNA samples were collected, and OsRDR1 mRNA levels relative to that of the mock-inoculated wild-type rice plant were determined by qRT-PCR. Results are means ± se for three replicates. Asterisks indicate significant differences between the wild-type plant and miR444-overexpressed lines (Student’s t test analysis, P < 0.05).
Dimerization of OsMADS23, OsMADS27a, and OsMADS57
Among of the four MADS box targets of miR444, OsMADS23, OsMADS27a, and OsMADS57 had typical MADS box domains. No obvious MADS box domain was identified for OsMADS27b, suggesting that OsMADS27b cannot regulate gene expression by binding directly to the gene promoter. Reverse transcription (RT)-PCR analysis showed that OsMADS23 and OsMADS27a were expressed in stems, leaves, roots, leaf sheaths, and shoots; OsMADS57 was expressed mainly in leaves, leaf sheaths, and shoots (Supplemental Fig. S1A). In subcellular localization assays, the fluorescence of the OsMADS23-, OsMADS27a-, and OsMADS57-GFP fusion proteins completely overlapped with 4′,6-diamidino-2-phenylindole nuclear staining in rice protoplasts (Supplemental Fig. S1B), indicating that the three miR444 target proteins are located in the nucleus.
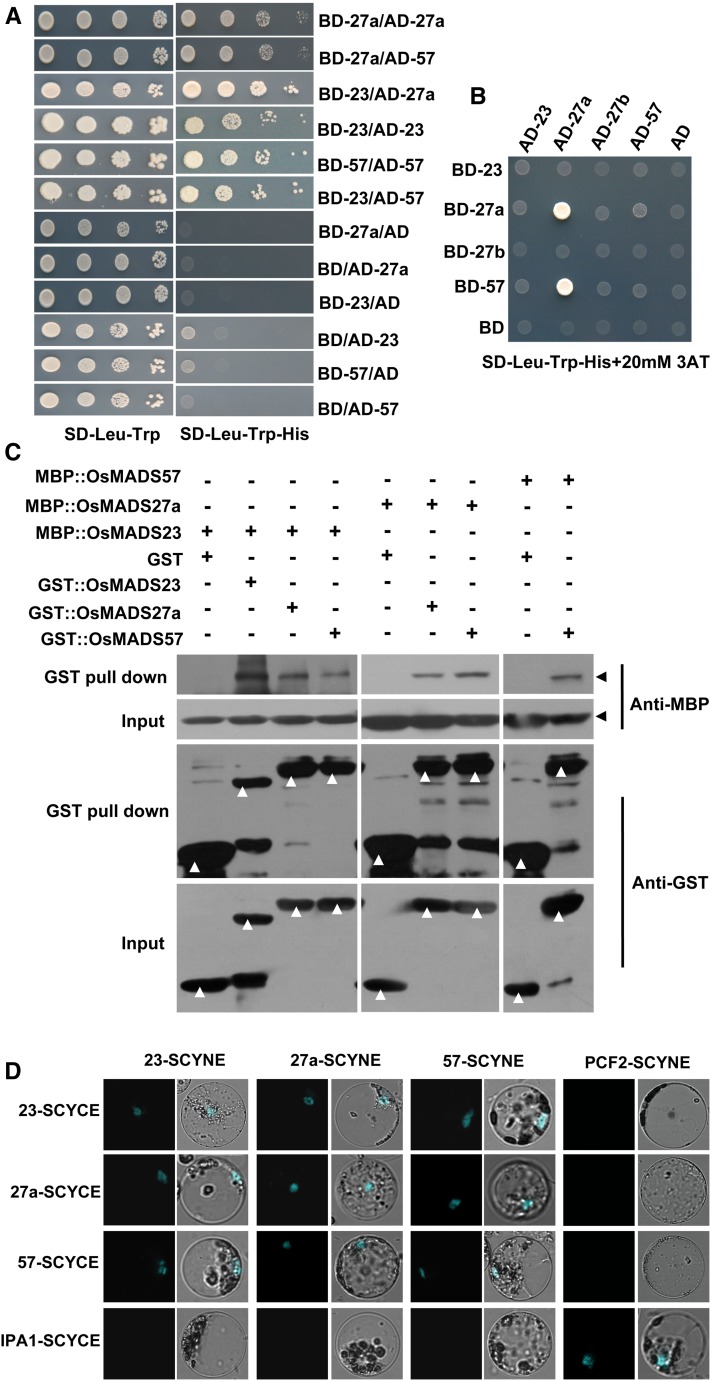
Yeast two-hybrid (Y2H) assays were performed to test the interactions of OsMADS23, OsMADS27a, and OsMADS57. To do this, OsMADS23, OsMADS27a, and OsMADS57 were fused to both the GAL activation domain and the GAL-binding domain. When the yeast (Saccharomyces cerevisiae) harboring different combinations grew in selective medium, we observed the interactions of OsMADS23, OsMADS27a, and OsMADS57 with themselves and with each other (Fig. 4A). Interestingly, very strong OsMADS27a homodimerization and OsMADS27a-OsMADS57 heterodimerization were observed when yeast grew on selective medium containing a high concentration of 3-amino-1,2,4-triazole (20 mm) in Y2H assays (Fig. 4B). OsMADS27b did not interact with OsMADS23, OsMADS27a, OsMADS57, or itself in yeast (Fig. 4B). Consistent with the observations in Y2H assays, pull-down assays also showed that OsMADS23, OsMADS27a, and OsMADS57 interacted with themselves and with each other (Fig. 4C). To confirm their interactions in planta, bimolecular fluorescence complementation (BiFC) assays were performed in rice protoplasts. Both N-terminal and C-terminal domains of cyan fluorescent protein (CFP) were fused to OsMADS23, OsMADS27a, and OsMADS57. OsMADS23-SCYNE and OsMADS23-SCYCE, OsMADS27a-SCYNE and OsMADS27a-SCYCE, and OsMADS57-SCYNE and OsMADS57-SCYCE were transiently coexpressed in rice protoplasts. The CFP fluorescence signal revealed the self-interactions of OsMADS23, OsMADS27a, and OsMADS57 (Fig. 4D). BiFC assays also were performed to examine the heterodimeric formation between OsMADS23, OsMADS27a, and OsMADS57. The results showed that OsMADS23, OsMADS27a, and OsMADS57 interacted with each other in rice protoplasts (Fig. 4D). Together, the above observations indicate that OsMADS23, OsMADS27a, and OsMADS57 form homodimers and heterodimers between them in rice.
Figure 4.
Dimerization of OsMADS23, OsMADS27a, and OsMADS57. A and B, Interaction patterns between OsMADS23, OsMADS27a, and OsMADS57 in Y2H assays. The yeast AH109 stain was cotransformed with the indicated constructs and grown on selective synthetic dropout (SD) medium. The construct was labeled by the gene name after AD (GAL activation domain) or BD (GAL-binding domain). 3AT, 3-Amino-1,2,4-triazole. C, Interaction patterns between OsMADS23, OsMADS27a, and OsMADS57 in GST pull-down assays. OsMADS23, OsMADS27a, and OsMADS57 were fused with GST and maltose-binding protein (MBP) tags and applied for GST pull-down assays. The interactions between OsMADS23, OsMADS27a, and OsMADS57 were detected by western blot using MBP antibody. Arrowheads indicate the expected bands of western blots. D, Interaction patterns between OsMADS23, OsMADS27a, and OsMADS57 in BiFC assays by cotransfecting rice protoplasts with the indicated constructs. Merged images of CFP fluorescence and rice protoplast are shown. The construct was labeled by the gene name followed by SCYNE (the N-terminal fragment of CFP) or SCYCE (the C-terminal fragment of CFP). The previously reported interacting proteins IPA1 (Ideal Plant Architecture1) and PCF2 (Proliferating cell nuclear antigen gene promoter binding factor 2) (Lu et al., 2013) were used as a positive control. The interactions between the three MADS proteins and IPA1 or PCF2 were detected and used as negative controls.
OsMADS23, OsMADS27a, and OsMADS57 Repress OsRDR1 Expression
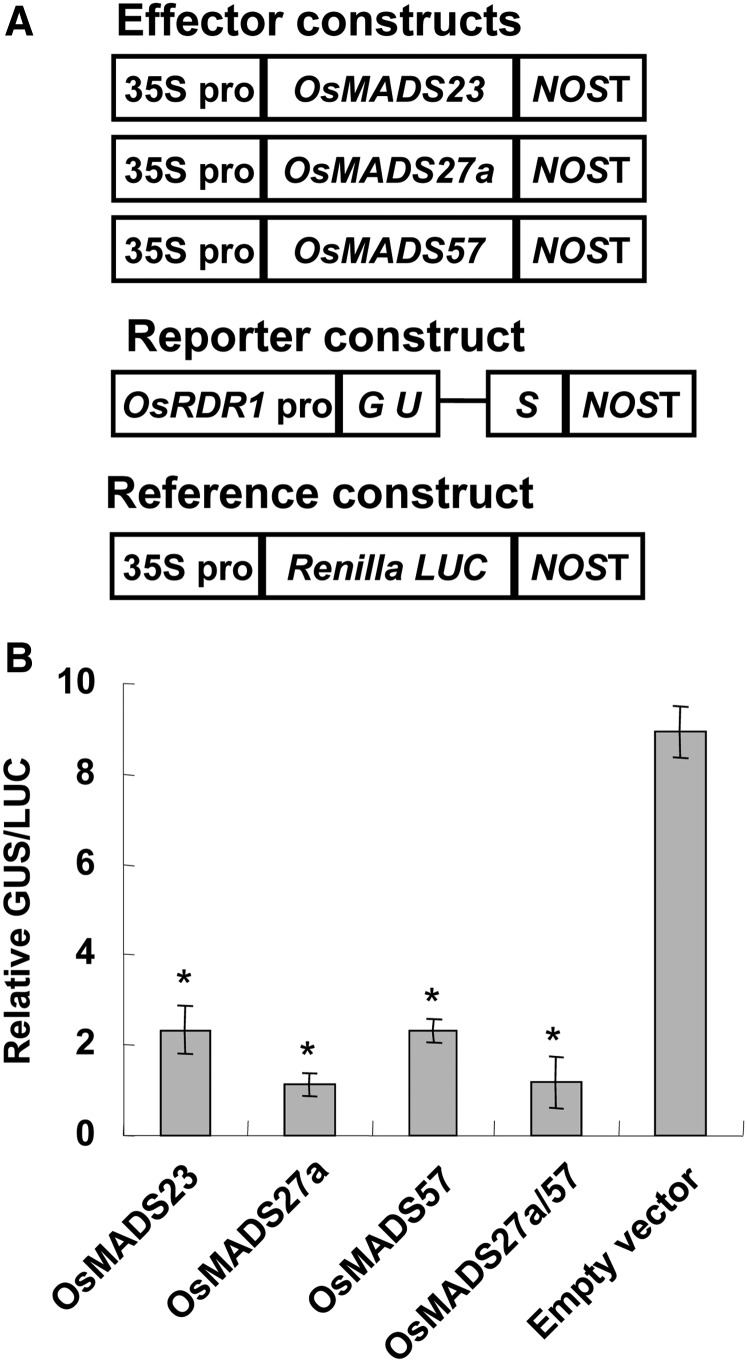
Interestingly, cis-element scanning of the approximately 2-kb promoter region showed that OsRDR1 had five typical CArG motifs (Supplemental Table S2). Recently, a transient expression assay in protoplasts showed that OsMADS57 is a transcriptional repressor (Guo et al., 2013), suggesting that OsMADS57 and its two interacting analogs, OsMADS23 and OsMADS27a, might repress OsRDR1 expression, and the miR444-mediated increase in OsRDR1 expression might be due to the attenuation of the repressive roles of OsMADS23, OsMADS27a, and OsMADS57 in OsRDR1 transcription. To verify this, we performed a transient expression assay. N. benthamiana leaves were coinfiltrated with Agrobacterium tumefaciens harboring different effector protein constructs (35Spro:OsMADS23, 35Spro:OsMADS27a, or 35Spro:OsMADS57) and the reporter construct (a GUS gene driven by the OsRDR1 promoter; Fig. 5A). GUS activity analysis revealed that OsMADS23, OsMADS27a, and OsMADS57 repressed the activity of the OsRDR1 promoter in coinoculated leaves (Fig. 5B). Comparably, OsMADS27a showed a higher repressive ability than OsMADS23 and OsMADS57 on GUS expression (Fig. 5B). This stronger repressive activity of OsMADS27a on the OsRDR1 promoter could be explained by the stronger interaction of OsMADS27a in homodimerization (Fig. 4B). Similarly, the strong interaction of OsMADS27a and OsMADS57 (Fig. 4B) may lead to a severe repression of OsRDR1 promoter activity when the GUS reporter construct is coinfiltrated with the 35Spro:OsMADS27a and 35Spro:OsMADS57 constructs (Fig. 5B). Together, these results indicate that OsMADS23, OsMADS27a, and OsMADS57 repress OsRDR1 expression. Reasonably, an increase in miR444 reduces the amounts of OsMADS23, OsMADS27a, and OsMADS57 (Fig. 1), thereby activating OsRDR1 expression to trigger the downstream cascade of the rice antiviral responses.
Figure 5.
OsMADS23, OsMADS27a, and OsMADS57 repress the expression of OsRDR1. A, Schematic structures of the effector and reporter constructs for the transient expression assay in N. benthamiana leaves, in which OsMADS23, OsMADS27a, and OsMADS57 were under the control of the Cauliflower mosaic virus (CaMV) 35S promoter, the GUS reporter gene harboring an intron was driven by the OsRDR1 promoter, and the Renilla luciferase (LUC; from Renilla reniformis) gene derived by the 35S promoter was used as an internal reference. B, Transient expression assay in N. benthamiana leaves. Relative GUS activities were normalized to the activities of Renilla luciferase and averaged from three biological repeats. Error bars indicate se. Asterisks indicate significant differences compared with the empty vector samples (Student’s t test analysis, P < 0.05).
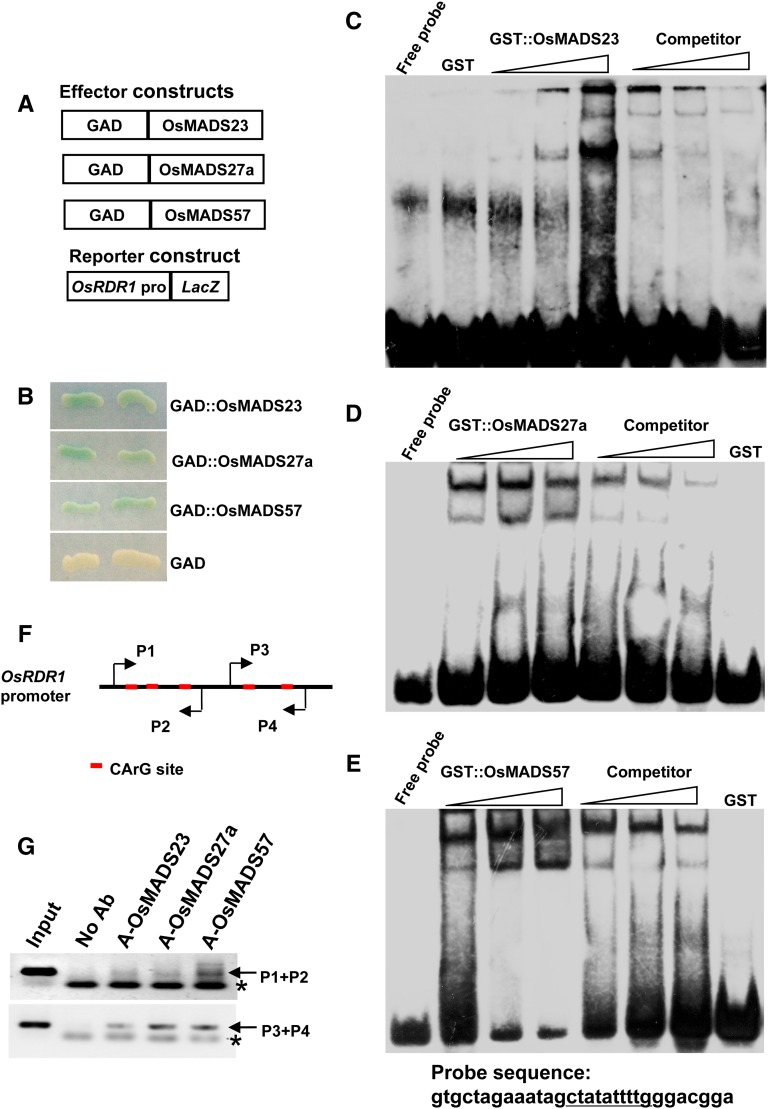
OsMADS23, OsMADS27a, and OsMADS57 Bind Directly to the CArG Motifs of the OsRDR1 Promoter in Planta
To examine if OsMADS23, OsMADS27a, and OsMADS57 repress the expression of OsRDR1 by binding directly to its promoter’s CArG motifs, three complementary approaches were employed. First, a yeast one-hybrid (Y1H) assay was performed by coexpression of the GAL4 activation domain (GAD)-fused OsMADS23, OsMADS27a, or OsMADS57 and the LacZ reporter gene driven by the OsRDR1 promoter fragment containing a CArG motif (Fig. 6A). The results showed that GAD-OsMADS23, GAD-OsMADS27a, and GAD-OsMADS57, but not GAD alone, activated the expression of the LacZ reporter gene (Fig. 6B). Second, to analyze protein-DNA complex formation, the OsMADS23, OsMADS27a, or OsMADS57 recombinant protein (fused to glutathione S-transferase [GST]) was incubated with a biotin-labeled CArG motif sequence or a CArG motif mutant sequence (the same as the CArG motif in the Y1H assay) of the OsRDR1 promoter, with or without competitor sequences, and then subjected to electrophoretic mobility shift assay (EMSA). Consistent with the observations in the Y1H assay, the gel-shift assays showed that the GST-OsMADS23, GST-OsMADS27a, and GST-OsMADS57 fusion proteins interacted with the CArG motif from the OsRDR1 promoter but not the CArG mutant sequence (Fig. 6, C–E; Supplemental Fig. S2). Finally, and more importantly, we confirm the binding of OsMADS23, OsMADS27a, and OsMADS57 to the OsRDR1 promoter in rice plants by chromatin immunoprecipitation (ChIP)-PCR assays. The results showed that OsMADS23, OsMADS27a, and OsMADS57 interacted with the OsRDR1 promoter regions harboring the CArG motifs (Fig. 6, F and G). Consistently, the repressive effects of OsMADS23, OsMADS27a, and OsMADS57 on the OsRDR1 promoter were abolished when the CArG motifs were deleted from the OsRDR1 promoter (Supplemental Fig. S3). Together, these observations indicate that OsMADS23, OsMADS27a, and OsMADS57 repress the expression of OsRDR1 by binding directly to the CArG motifs of its promoter in planta.
Figure 6.
OsMADS23, OsMADS27a, and OsMADS57 bind to the CArG motifs of the OsRDR1 promoter. A, Schematic structures of the effector and reporter constructs for the Y1H assay. OsMADS23, OsMADS27a, and OsMADS57 were fused to the GAD, and the reporter gene LacZ was driven by the OsRDR1 promoter fragment containing a CArG motif. B, OsMADS23, OsMADS27a, and OsMADS57 bound to the promoter fragment of OsRDR1 in yeast. C to E, EMSAs showed that OsMADS23, OsMADS27a, and OsMADS57 bound to the CArG motif of the OsRDR1 promoter. The biotinylated probe containing the CArG motif sequence was incubated with GST-OsMADS23, GST-OsMADS27a, or GST-OsMADS57, while the probe incubated with no protein or GST protein was used as a negative control. Nonlabeled probes were used as cold competitors. F and G, ChIP-PCR assays showed that OsMADS23, OsMADS27a, and OsMADS57 bound to the OsRDR1 promoter regions containing CArG motifs (red boxes) in rice plants. Two pairs of PCR primers (P1/P2 and P3/P4) are indicated, and the amplified PCR bands are indicated by arrows. Asterisks indicate the dimers formed by primers. Ab, Antibody.
Overexpression of miR444-Resistant OsMADS57 Reduces OsRDR1 Expression and Rice Resistance against Virus Infection
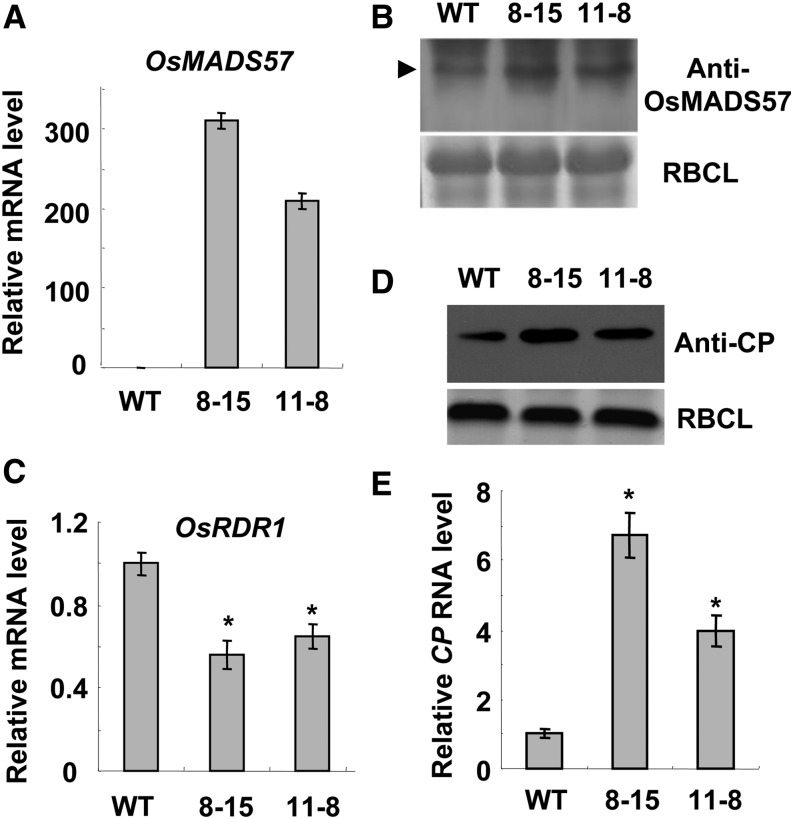
In order to further illustrate the repressive roles of OsMADS23, OsMADS27a, and OsMADS57 on OsRDR1 transcription during virus infection, we attempted to overexpress miR444-resistant OsMADS23, OsMADS27a, and OsMADS57 (named OsMADS23R, OsMADS27aR, and OsMADS57R) in rice plants to see if they can inhibit OsRDR1 expression and damage the virus resistance. No OsMADS23R- and OsMADS27aR-overexpressed rice seeds could be obtained, suggesting that the overexpression of OsMADS23 and OsMADS27a was detrimental for embryo development or seed germination, like the effect of ectopic expression of their orthologous gene AtANR1 in Arabidopsis (Gan et al., 2012). Rice plants overexpressing OsMADS57R were generated normally with greatly increased expression levels of OsMADS57R and OsMADS57 protein (Fig. 7, A and B). These rice plants were challenged with RSV. At 14 dpi, the levels of OsRDR1 mRNA, the RSV CP, and its RNA in plants were analyzed. The results showed that OsMADS57R-overexpressed rice plants showed lower OsRDR1 mRNA levels compared with wild-type rice plants during virus infection (Fig. 7C). Consistently, OsMADS57R-overexpressed rice plants accumulated much higher levels of the RSV CP protein and RNA than wild-type rice plants (Fig. 7, D and E). Together, these results indicate that overexpression of OsMADS57R repressed the expression of OsRDR1, resulting in a rice more susceptible to RSV infection.
Figure 7.
Overexpression of miR444-resistant OsMADS57 (named OsMADS57R) reduces OsRDR1 expression and rice resistance against RSV infection. A, Analysis of the mRNA levels of OsMADS57R by qRT-PCR in transgenic rice lines (8-15 and 11-8) overexpressing miR444-resistant OsMADS57. B, Analysis of the accumulation of OsMADS57 protein by western blot in transgenic rice lines (8-15 and 11-8) overexpressing miR444-resistant OsMADS57. The arrowhead indicates the bands of OsMADS57. C, Analysis of the expression levels of OsRDR1 by qRT-PCR in OsMADS57R-overexpressed rice plants relative to that of wild-type (WT) rice plants at 14 dpi of RSV infection. D and E, Analysis of the accumulation of RSV CP by western blot and the RSV CP RNA by qRT-PCR in OsMADS57R-overexpressed and wild-type rice plants at 14 dpi of RSV infection. The Rubisco large subunit (RBCL) bands were visualized by Coomassie Brilliant Blue and served as a loading control in western blots. The qRT-PCR results are means ± se for three replicates. Asterisks indicate significant differences between wild-type and OsMADS57R-overexpressed rice plants (Student’s t test analysis, P < 0.05).
Knockout of OsRDR1 Reduces Rice Resistance against Virus Infection
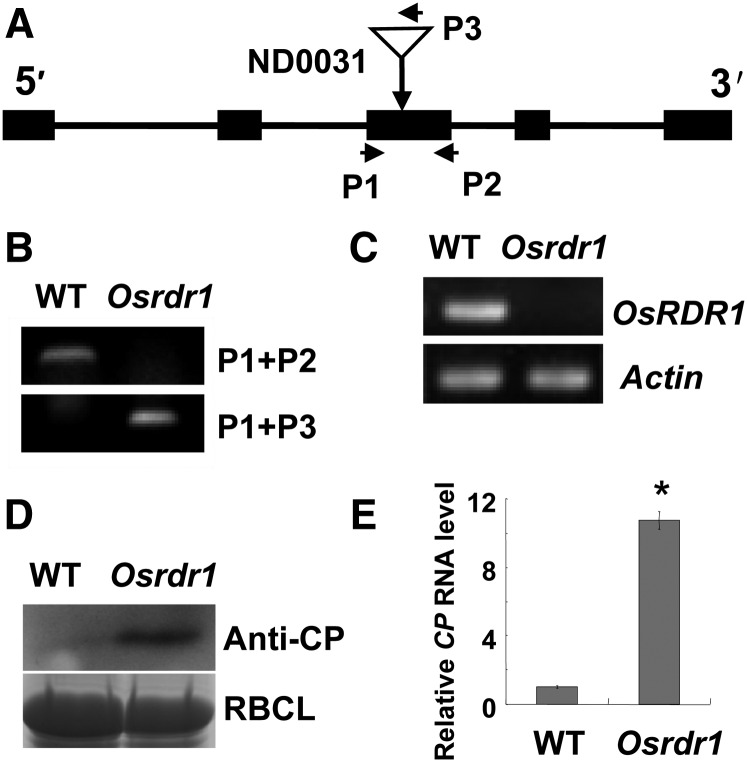
Having elucidated the signaling steps in the rice antiviral pathway from RSV infection to OsRDR1 expression through the actions of miR444 and its target proteins, we further sought evidence to support a positive role of OsRDR1 in rice virus resistance. In a loss-of-function experiment, an Osrdr1 mutant with a Tos17 inserted in the third exon of the OsRDR1 gene (obtained from the Tos17 insertion mutant library; accession no. ND0031) was used (Fig. 8, A and B). Semiquantitative RT-PCR analysis using gene-specific primers verified that Osrdr1 completely lost the expression of OsRDR1 (Fig. 8C). To determine the role of OsRDR1 in the rice antiviral response, Osrdr1 and wild-type rice plants were grown in soil, and their seedlings were challenged with RSV. Then, the levels of the RSV CP and its RNA in plants, indicative of viral propagation, were analyzed. The results showed that Osrdr1 accumulated much higher levels of the RSV CP protein and RNA than wild-type rice plants (Fig. 8, D and E), indicating that OsRDR1 plays important roles in rice virus resistance.
Figure 8.
Knockout of OsRDR1 reduces rice resistance against RSV infection. A, Schematic structure of the Tos17 insertion site of the Osrdr1 mutant (ND0031). B, Homozygous Osrdr1 was verified by PCR assays using OsRDR1-specific primers (P1+P2) and OsRDR1- and Tos17-specific primers (P1+P3). C, RT-PCR analysis using gene-specific primers showing that the expression of OsRDR1 was knocked out in the Osrdr1 mutant. D, Analysis of the accumulation of RSV CP by western blot in Osrdr1 mutant and wild-type (WT) rice plants at 14 dpi of RSV infection. The Rubisco large subunit (RBCL) bands were visualized by Coomassie Brilliant Blue and served as a loading control. E, Analysis of the levels of the RSV CP RNA by qRT-PCR in the Osrdr1 mutant relative to that of wild-type rice plants at 14 dpi of RSV infection. Results are means ± se for three replicates. The asterisk indicates a significant difference between wild-type and Osrdr1 rice plants (Student’s t test analysis, P < 0.05).
DISCUSSION
miR444 Positively Regulates the Rice Antiviral Response
miRNAs play important regulatory functions in plant immunity against diverse bacterial, fungal, and viral pathogens (Ruiz-Ferrer and Voinnet, 2009; Katiyar-Agarwal and Jin, 2010; Weiberg et al., 2014). Here, we showed that miR444 accumulation was induced by RSV infection (Fig. 1A). Overexpression of miR444 increased rice resistance against RSV infection (Fig. 2), indicating that miR444 positively regulated the rice antiviral response. One of the possible mechanisms of miR444-mediated virus resistance is the activation of the expression of RDR1 (Fig. 3), which has been implicated in antiviral resistance via several pathways (Diaz-Pendon et al., 2007; Qi et al., 2009; Garcia-Ruiz et al., 2010; Wang et al., 2010; Cao et al., 2014). In fact, we have shown in this study that OsRDR1 was required for rice resistance against RSV infection (Fig. 8). Besides, our RNA-seq data showed that miR444 also up-regulated the expression of several R genes (Supplemental Table S1), suggesting that miR444 may play roles in R gene-mediated immunity in rice. Consistently, RSV-insusceptible rice plants (japonica cv Zhendao88) showed markedly elevated miR444 levels after RSV infection (Supplemental Fig. S4). miR444 might regulate the expression of these R genes also through its MADS box targets, because the promoters of these R genes contained CArG motifs (data not shown) as in the OsRDR1 promoter.
It has been reported that miR444 accumulation was up-regulated in rice also by bacterial and fungal infection (Li et al., 2014; http://mpss.udel.edu/#rice). But the roles that miR444 played during these microbial infections are currently unknown. As miR444 already has been reported to regulate rice tillering and nitrate signaling (Guo et al., 2013; Yan et al., 2014), we now add a new role to miR444, that of a regulator in the rice antiviral response.
miR444-Regulated Nitrate Signaling May Play Roles in the Rice Antiviral Response
Regulatory roles of miR444 in both rice nitrate signaling and antiviral response (Yan et al., 2014; Fig. 1) prompt us to wonder if miR444-regulated nitrate signaling plays roles in the rice antiviral response, because nitrogen is an important factor in plant disease resistance, besides its roles in plant growth and development. For example, nitrogen-rich fertilizer can reduce the incidence of the fungal pathogen take-all (Gaeumannomyces graminis) in barley (Hordeum vulgare) and wheat (Triticum aestivum; Huber and Haneklaus, 2007). In some situations, improved nitrogen nutrition can reduce or even eliminate viral disease symptoms (Spann and Schumann, 2010). Previous studies have suggested several mechanisms by which nitrogen affects plant disease resistance. First, although immunity is genetically controlled in plants, adequate nutritional status is important for the full expression of disease resistance. Second, nitrogen is essential to the generation of various cellular structures, proteins, and enzymes that are needed to develop defensive cell walls and produce antimicrobial compounds, such as phytoalexins, antioxidants, and flavonoids (Spann and Schumann, 2010). Third, nitrogen-containing metabolites can directly regulate the plant immunity signaling pathway. For instance, nitric oxide as a signal molecule is required for the hypersensitive reaction (Delledonne et al., 1998). Alteration of amino acid homeostasis in lht1 (for Lys and His transport) modulates salicylic acid-associated redox status and defense responses to Pseudomonas syringae (Liu et al., 2010).
miR444-regulated nitrate signaling may play roles in improving rice resistance to virus potentially by manipulating the nitrogen status during virus infection. On the one hand, since overexpression of miR444 increased nitrate accumulation (Yan et al., 2014), increased miR444 accumulation during RSV infection (Fig. 1; Supplemental Fig. S4) might provide rice with adequate nitrogen nutrition for disease resistance. On the other hand, as RSV first infects the newly emerged rice leaves, overexpression of miR444 would disrupt the remobilization of nitrogen from old leaves to young leaves (Yan et al., 2014), leaving an unfavorable nitrogen nutrition condition for virus multiplication. Future studies need to test whether the accumulation of miR444 could be augmented by the nutritional status during virus infection so that it can defend rice against virus infection by simultaneously regulating nitrogen content/distribution and activating defense genes, including OsRDR1.
miR444 Activates the OsRDR1-Dependent Antiviral RNA Silencing Pathway by Targeting MIKCC-Type MADS Box Proteins
RNA silencing-based antiviral activity plays a key role in host defense against virus infection (Ding and Voinnet, 2007; Ding, 2010; Pumplin and Voinnet, 2013). The expression of the key factors of RNA-silencing pathways usually is activated by virus infection at the transcriptional level to increase the activities of antiviral RNA silencing. For example, the expression of AGO1, AGO2, or RDR1 was usually induced by infection with diverse viruses (Yang et al., 2004; Alamillo et al., 2006; Zhang et al., 2006; Azevedo et al., 2010; He et al., 2010; Satoh et al., 2010; Várallyay et al., 2010; Du et al., 2011; Harvey et al., 2011). However, the molecular mechanisms of activation of the expression of these antiviral genes are unknown. In this study, we demonstrated a signaling cascade of activation of RDR1 expression in the rice antiviral response. We showed that an increase in miR444 accumulation improved the rice antiviral response by up-regulation of the expression of OsRDR1 (Figs. 1–3). We further showed that three miR444 targets (i.e. the MIKCC-type MADS box proteins OsMADS23, OsMADS27a, and OsMADS57) bound directly to the CArG motifs of the OsRDR1 promoter and repressed OsRDR1 expression (Figs. 5 and 6). Thus, increased levels of miR444 diminished the repressive roles of OsMADS23, OsMADS27a, and OsMADS57 on OsRDR1 transcription, leading to the activation of the OsRDR1-dependent antiviral pathway upon virus infection. Consistently, overexpression of miR444-resistant OsMADS57 reduces OsRDR1 expression and results in rice more susceptible to virus infection (Fig. 7). OsMADS23, OsMADS27a, and OsMADS57 could form homodimers and heterodimers between them (Fig. 4), suggesting that these protein dimers or even their multimeric complexes control the expression of OsRDR1. Previous reports showed that multiple key components of the antiviral RNA-silencing pathway, like DCL1, AGO1, and AGO2, were negatively regulated by their cognate miRNAs at the posttranscriptional level (Bologna and Voinnet, 2014). Our study revealed that RDR1, another key component of the antiviral RNA-silencing pathway, is regulated by miR444 at the transcriptional level through its target proteins, adding a new regulatory mechanism controlling the activity of antiviral RNA silencing. As far as we know, this is the first demonstration that an miRNA evolves its antiviral function and is involved in the activation of the antiviral RNA-silencing pathway. miR444 is a monocot-specific miRNA. Whether other monocot plants also exploit a similar signaling cascade to activate the RDR1-dependent antiviral RNA-silencing pathway needs to be investigated further. A bioinformatics search indicated that wheat and Brachypodium distachyon have equivalents of miR444 and its target proteins, CArG motif-containing RDR1 genes, indicating that this might be the case. Comparably, the dicot plants only have homologs of miR444’s target proteins, such as ANR1 in Arabidopsis (Arora et al., 2007). The AtRDR1 promoter also contains a CArG motif. It will be interesting to see if the homolog proteins of the miR444 targets regulate the expression of RDR1 of dicot plants in response to virus infection.
Besides directly silencing viral RNAs, RNA-silencing pathways also potentially regulate host genes to build virus resistance. Recently, it was reported that RDR1 played a role in activating broad-spectrum antiviral activity by the production of a distinct class of virus-activated endogenous siRNAs to direct the widespread silencing of host genes in Arabidopsis (Cao et al., 2014), indicating that RDR1 performed the antiviral roles by silencing both viral RNAs and host immunity-related genes. Interestingly, Our RNA-seq data showed that miR444 regulated the expression of hundreds of rice genes related to the plant-pathogen interaction (Supplemental Table S1). A potential mechanism was that the miR444-activated OsRDR1-dependent RNA-silencing pathway regulated rice endogenous genes involved in broad-spectrum immunity, as shown in Arabidopsis. However, we could not exclude the possibility that the miR444 targets regulated these immunity-related genes in other OsRDR1-independent ways. A future study using Osrdr1 mutant and OsRDR1-overexpressed rice plants will be helpful to elucidate the miR444-mediated antiviral mechanisms.
CONCLUSION
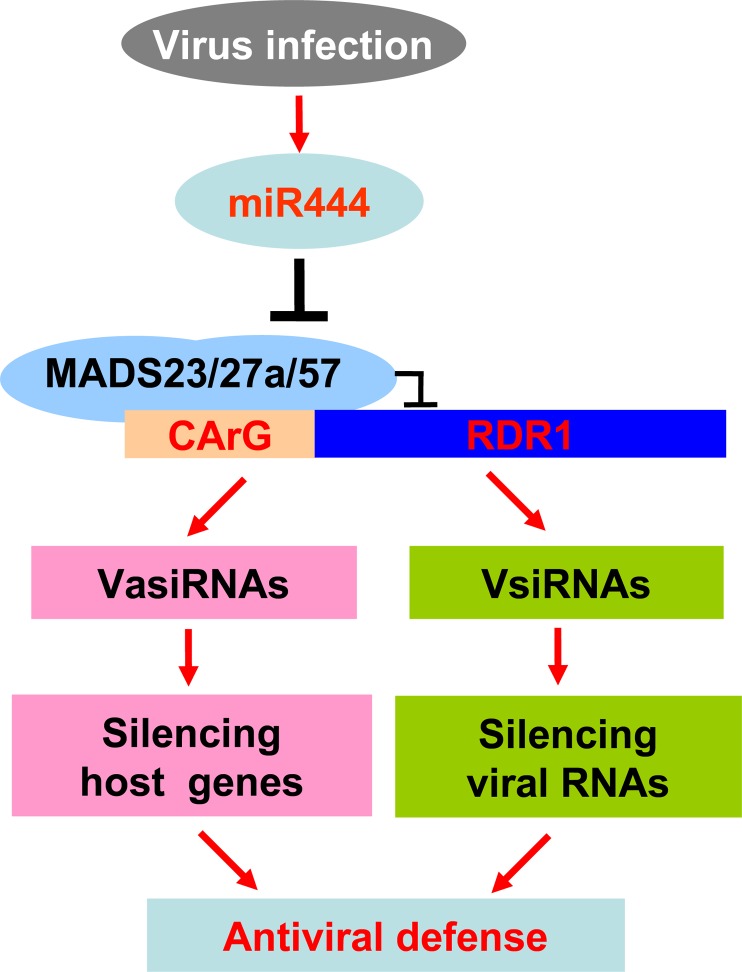
In conclusion, our studies reveal a novel signaling cascade in the rice antiviral RNA-silencing pathway (Fig. 9). Without virus infection, the expression of OsRDR1 is repressed by OsMADS23, OsMADS27a, and OsMADS57. Upon virus infection, miR444 is induced to diminish the expression of OsMADS23, OsMADS27a, and OsMADS57. Then, the OsRDR1-dependent RNA-silencing pathway is activated to defend rice against virus infection by silencing both viral RNAs and host genes. Our study also points to a role of miR444-regulated nitrate signaling in subsidizing the rice antiviral response, presumably by manipulating the nitrogen status, suggesting a function of miR444 in the cross talk of nitrate signaling and the antiviral response.
Figure 9.
Model of the miR444-RDR1 signaling cascade in the rice antiviral RNA-silencing pathway. Three miR444 targets, OsMADS23, OsMADS27a, and OsMADS57, form homodimers, heterodimers, or even polymers between them to repress the expression of OsRDR1 by binding directly to the CArG motifs of its promoter. Upon virus infection, miR444 expression is induced. Consequently, an increased level of miR444 diminishes the repressive roles of OsMADS23, OsMADS27a, and OsMADS57 on OsRDR1 transcription. Then, the OsRDR1-dependent RNA-silencing pathway is activated to defend against viral infection by producing vsiRNAs to directly silence viral RNAs and virus-activated siRNAs (VasiRNAs) to silence host genes for the activation of broad-spectrum antiviral activity.
MATERIALS AND METHODS
Plant Growth and RSV Infection
RSV-free and viruliferous insects (Laodelphax striatellus) were obtained and maintained as described previously (Huo et al., 2014). RSV inoculation was performed as described previously (Satoh et al., 2010). Briefly, rice (Oryza sativa) seeds were grown in a pot at 25°C ± 3°C with a 16-h-light/8-h-dark photoperiod and approximately 70% humidity in a growth incubator. After 2 weeks, 16 rice seedlings at the two-leaf stage in a single pot were exposed to virus-free (for inoculation control) or viruliferous insects, with a plant:insect ratio of 1:3. After 2 d of feeding, the insects were removed and the seedlings continued to grow for sample collection and observation of disease symptoms.
Plasmid Constructs
To detect the subcellular localizations of OsMADS23, OsMADS27a, and OsMADS57 in rice protoplast, the full-length complementary DNA (cDNA) sequences of OsMADS23, OsMADS27a, and OsMADS57 were amplified and cloned separately into pBI121 vector upstream of the GFP gene under the control of the CaMV 35S promoter. To express OsMADS23, OsMADS27a, and OsMADS57 transiently in Nicotiana benthamiana, OsMADS23, OsMADS27a, and OsMADS57 were cloned separately into pCAMBIA1300 vector downstream of the CaMV 35S promoter. To generate the OsRDR1 promoter-GUS fusion construct, an approximately 2-kb promoter sequence of OsRDR1 was amplified and cloned into pBI121 vector upstream of the GUS gene harboring an intron. To generate the CArG-deleted OsRDR1 promoter-GUS fusion construct, an approximately 1.3-kb sequence of OsRDR1 promoter containing all five CArG motifs was deleted. The approximately 700-bp promoter sequence immediately upstream the 5′ untranslated region of OsRDR1 was amplified and cloned into pBI121 vector upstream of the GUS gene harboring an intron. For BiFC assays, OsMADS23, OsMADS27a, and OsMADS57 were cloned separately into pSCYCE and pSCYNE(R) (Waadt et al., 2008). For Y1H assays, OsMADS23, OsMADS27a, and OsMADS57 were cloned separately into pGAD424 vector. The truncated promoter sequence harboring the CArG motif of OsRDR1 was cloned into pLacZi2μ vector. For Y2H assays, OsMADS23, OsMADS27a, and OsMADS57 were cloned separately into pDEST22 and pDEST32 vectors (Invitrogen) from the cognate pDonor 221 constructs by recombination. For the expression of recombinant proteins of OsMADS23, OsMADS27a, and OsMADS57 in Escherichia coli, OsMADS23, OsMADS27a, and OsMADS57 were cloned separately into pET30a vector downstream of the GST gene and the MBP gene. For the transformation of miR444-resistant OsMADS23, OsMADS27a, and OsMADS57 to rice plants, OsMADS23, OsMADS27a, and OsMADS57 were mutated to miR444 cleavage-resistant versions with unaltering amino acid sequences. Then, miR444-resistant OsMADS23, OsMADS27a, and OsMADS57 were cloned separately into pCAMBIA1300 vector downstream of the CaMV 35S promoter. Primers are listed in Supplemental Table S3.
Small RNA Gel-Blot Analysis
Small RNA gel-blot analysis was performed as described previously (Yan et al., 2011). Briefly, total RNA was extracted from rice samples with Trizol reagent (Invitrogen). RNA samples were separated on a denaturing 17% (w/v) polyacrylamide gel and transferred electrophoretically to Hybond-N+ membranes (Amersham Bioscience). Hybridizations were performed at 38°C in PerfectHyb Plus buffer with DNA oligonucleotide probes 32P labeled by T4 polynucleotide kinase (New England Biolabs). Hybridization signals were detected with a phosphorimager. Sequences of the probes are listed in Supplemental Table S3.
qRT-PCR Analysis
DNA-free total RNA (3 μg) was subjected to RT using SuperScript III reverse transcriptase (Invitrogen) to produce cDNA products following the supplier’s protocol. qRT-PCR was performed by adding SYBR Green real-time PCR master mix (Toyobo) to the reaction system. Three replicates were performed for each gene. The relative quantification of each sample was determined by normalization against the amount of GLYCERALDEHYDE 3-PHOSPHATE cDNA detected in the same sample. Primers are listed in Supplemental Table S3.
Western-Blot Analysis
Rice samples were ground in liquid nitrogen and homogenized in protein extraction buffer containing 140 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, and 1.8 mm KH2PO4. Total protein was separated on an SDS-PAGE gel and transferred electrophoretically to polyvinylidene difluoride membranes (Amersham Bioscience), which were incubated with purified polyclonal antibodies (anti-OsMADS23, anti-OsMADS27a, anti-OsMADS57 [BGI], and anti-CP [Huo et al., 2014]) to determine protein accumulation.
Pull-Down Assays
For in vivo pull-down experiments, total proteins were extracted from E. coli strain BL21 expressing GST- or MBP-fused OsMADS23, OsMADS27a, or OsMADS57 using STE buffer (10 mm Tris-HCl [pH 8], 1 mm EDTA [pH 8], and 150 mm NaCl) or column buffer (20 mm Tris-HCl [pH 7.4], 1 mm EDTA [pH 8], and 200 mm NaCl), respectively. Then, mixtures of different total proteins were incubated with glutathione-Sepharose beads (GE). After being centrifuged and washed, the protein-bound beads were loaded on an SDS-PAGE gel to perform western-blot assays using antibodies to MBP and GST.
Subcellular Localization and BiFC Assays
Rice protoplast isolation and transfection were performed as described previously (Zhang et al., 2011). For subcellular localization and BiFC assays, protoplasts were transfected with the indicated plasmids or plasmid mixtures by a polyethylene glycol-mediated method. After approximately 12 h of incubation in the dark, GFP or CFP fluorescence was observed with a Leica SP8 microscope.
Transient Expression in N. benthamiana
The transient expression assays were performed as described previously (Liu et al., 2003). Briefly, Agrobacterium tumefaciens (GV3101) transformants harboring the designated constructs were grown overnight in a culture with 50 μg mL−1 kanamycin, 10 mm MES, and 20 μm acetosyringone. Agrobacterial cells were harvested by centrifugation and resuspended in MMA buffer (10 mm MgCl2, 10 mm MES [pH 5.6], and 100 μm acetosyringone) to an optical density at 600 nm of 1. To examine the repressive activities of OsMADS23, OsMADS27a, and OsMADS57 on OsRDR1 transcription, agrobacterial cells harboring the designated reporter construct (OsRDR1 pro:GUS or CArG-deleted OsRDR1 pro:GUS), an effector construct (35S:OsMADS23, 35S:OsMADS27a, 35S:OsMADS57, or vector), and an internal control construct (35S:Riluc) were mixed with a ratio of 2:3:1. After incubation at room temperature for 3 h, the agrobacterial cell suspension containing different construct combinations was pressure infiltrated into N. benthamiana leaves. The leaves were harvested using hole punch 2 d after infiltration. For analysis of GUS and Renilla luciferase activities, total protein was extracted using passive lysis buffer (Promega). GUS activities were detected as described previously (Liu et al., 2003). The Renilla luciferase activities were analyzed using the Renilla Luciferase Assay System Kit (Promega) following the supplier’s protocol. The repressive activities of OsMADS23, OsMADS27a, and OsMADS57 were determined as the relative ratio of GUS to Renilla luciferase activity.
EMSAs
EMSAs were performed using the Light Shift Chemiluminescent EMSA Kit following the supplier’s protocol (Thermo Scientific). Briefly, 2 nm biotin-labeled probes and purified fusion proteins with gradient concentrations (0.2, 2.5, and 5 pm) were incubated in a 20-μL reaction mixture at room temperature for 30 min. Then, the reaction mixtures were separated on a 6% native polyacrylamide gel and transferred electrophoretically to Hybond-N+ membranes (Amersham Bioscience). For the competition assays, unlabeled probe sequences (5-, 10-, or 50-fold of the labeled probe sequences) were added to the EMSA reactions for GST::OsMADS23, and unlabeled probe sequences (100-, 200-, or 400-fold of the labeled probe sequences) were added to the EMSA reactions for GST::OsMADS27a and GST::OsMADS57. Labeled fragments and their shifted complexes with proteins were visualized using the Chemiluminescent Nucleic Acid Detection Module Kit (Thermo Scientific). The probe sequence for OsRDR1 (or mutant) was 5′-GTGCTAGAAATAGCTATATTTTGGGACGGA-3′ or 5′-GTGCTAGAAATAGAAAAAAAAAAGGACGGA-3′, in which the CArG (or mutant) motif sequence is underlined.
Y1H Assays
Plasmids for the GAD fusion effector protein (pGAD424-OsMADS23, pGAD424-OsMADS27a, or pGAD424-OsMADS57) and the LacZ reporter gene driven by the OsRDR1 promoter fragment were cotransformed into Saccharomyces cerevisiae strain EGY48. The transformants were grown on proper drop-out plates (lacking Ura and Leu) containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside for blue color selection.
Y2H Assays
For Y2H assays, the designated plasmids pDEST-32 and pDEST-22 harboring OsMADS23, OsMADS27a, and OsMADS57 were mixed and transformed into S. cerevisiae strain AH109. The transformed yeast strains containing different construct combinations were plated on medium lacking Leu and Trp at 28°C for 2 d. To analyze the interactions, yeast transformants were screened by growing on selective medium lacking Leu, Trp, and His plus 20 mm 3-amino-1,2,4-triazole.
ChIP-PCR
ChIP-PCR assays were performed according to the method described previously (Saleh et al., 2008). Briefly, 2-week-old shoots (3 g) of wild-type plants were harvested and ground to powder in liquid nitrogen after cross-linking with 1% (v/v) formaldehyde under vacuum for 8 min. Then, the chromatin complexes were isolated and sonicated for ChIP by incubating with purified polyclonal antibodies (anti-OsMADS23, anti-OsMADS27a, and anti-OsMADS57 [BGI]). The precipitated DNA was recovered and dissolved in water as the templates for PCR analysis.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Expression patterns and protein localizations of OsMADS23, OsMADS27a, and OsMADS57.
Supplemental Figure S2. EMSAs showed that OsMADS23, OsMADS27a, and OsMADS57 bound to the CArG motif of the OsRDR1 promoter but not the CArG mutant sequence.
Supplemental Figure S3. OsMADS23, OsMADS27a, and OsMADS57 repress the expression of OsRDR1 through binding to the CArG motifs.
Supplemental Figure S4. miR444 accumulation was induced by virus infection in RSV-insusceptible rice plants.
Supplemental Table S1. Differentially expressed genes in both 2-1 and 13-1 lines.
Supplemental Table S2. Summary of the CArG motifs of the approximately 2-kb region of the OsRDR1 promoter.
Supplemental Table S3. Probes and primers used in this study.
Supplementary Material
Acknowledgments
We thank Kang Chong (Institute of Botany, Chinese Academy of Sciences) for providing the plasmids pGAD424 and pLacZi2μ and yeast strain EGY48 and Jiayang Li (Institute of Genetic and Developmental Biology, Chinese Academy of Sciences) for assisting with the BiFC system.
Glossary
- miRNA
microRNA
- siRNA
small interfering RNA
- dsRNA
double-stranded RNA
- vsiRNA
virus-derived small interfering RNA
- NBS-LRR
nucleotide-binding site-leucine-rich repeat
- RSV
Rice stripe virus
- dpi
days post inoculation
- RNA-seq
RNA sequencing
- qRT
quantitative reverse transcription
- Y2H
yeast two-hybrid
- BiFC
bimolecular fluorescence complementation
- Y1H
yeast one-hybrid
- EMSA
electrophoretic mobility shift assay
- ChIP
chromatin immunoprecipitation
- cDNA
complementary DNA
- CaMV
cauliflower mosaic virus
Footnotes
This work was supported by the National Basic Research Program of China (grant no. 2013CBA01403), the National Natural Science Foundation of China (grants nos. 31123007 and 31101424), and the Youth Innovation Promotion Association Foundation of the Chinese Academy of Sciences (grant no. Y52R012CR1).
References
- Alamillo JM, Saénz P, García JA (2006) Salicylic acid-mediated and RNA-silencing defense mechanisms cooperate in the restriction of systemic spread of plum pox virus in tobacco. Plant J 48: 217–227 [DOI] [PubMed] [Google Scholar]
- Arora R, Agarwal P, Ray S, Singh AK, Singh VP, Tyagi AK, Kapoor S (2007) MADS-box gene family in rice: genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genomics 8: 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ. (2013) Classification and comparison of small RNAs from plants. Annu Rev Plant Biol 64: 137–159 [DOI] [PubMed] [Google Scholar]
- Azevedo J, Garcia D, Pontier D, Ohnesorge S, Yu A, Garcia S, Braun L, Bergdoll M, Hakimi MA, Lagrange T, et al. (2010) Argonaute quenching and global changes in Dicer homeostasis caused by a pathogen-encoded GW repeat protein. Genes Dev 24: 904–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bologna NG, Voinnet O (2014) The diversity, biogenesis, and activities of endogenous silencing small RNAs in Arabidopsis. Annu Rev Plant Biol 65: 473–503 [DOI] [PubMed] [Google Scholar]
- Cao M, Du P, Wang X, Yu YQ, Qiu YH, Li W, Gal-On A, Zhou C, Li Y, Ding SW (2014) Virus infection triggers widespread silencing of host genes by a distinct class of endogenous siRNAs in Arabidopsis. Proc Natl Acad Sci USA 111: 14613–14618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Tamai A, Mori M, Ugaki M, Tanaka Y, Samadder PP, Miyao A, Hirochika H, Yamaoka N, Nishiguchi M (2010) Analysis of rice RNA-dependent RNA polymerase 1 (OsRDR1) in virus-mediated RNA silencing after particle bombardment. J Gen Plant Pathol 76: 152–160 [Google Scholar]
- de Folter S, Angenent GC (2006) trans meets cis in MADS science. Trends Plant Sci 11: 224–231 [DOI] [PubMed] [Google Scholar]
- Delledonne M, Xia Y, Dixon RA, Lamb C (1998) Nitric oxide functions as a signal in plant disease resistance. Nature 394: 585–588 [DOI] [PubMed] [Google Scholar]
- Diaz-Pendon JA, Li F, Li WX, Ding SW (2007) Suppression of antiviral silencing by cucumber mosaic virus 2b protein in Arabidopsis is associated with drastically reduced accumulation of three classes of viral small interfering RNAs. Plant Cell 19: 2053–2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding SW. (2010) RNA-based antiviral immunity. Nat Rev Immunol 10: 632–644 [DOI] [PubMed] [Google Scholar]
- Ding SW, Voinnet O (2007) Antiviral immunity directed by small RNAs. Cell 130: 413–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du P, Wu J, Zhang J, Zhao S, Zheng H, Gao G, Wei L, Li Y (2011) Viral infection induces expression of novel phased microRNAs from conserved cellular microRNA precursors. PLoS Pathog 7: e1002176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa M, Nakano T, Shima Y, Ito Y (2013) A large-scale identification of direct targets of the tomato MADS box transcription factor RIPENING INHIBITOR reveals the regulation of fruit ripening. Plant Cell 25: 371–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan Y, Bernreiter A, Filleur S, Abram B, Forde BG (2012) Overexpressing the ANR1 MADS-box gene in transgenic plants provides new insights into its role in the nitrate regulation of root development. Plant Cell Physiol 53: 1003–1016 [DOI] [PubMed] [Google Scholar]
- Garcia-Ruiz H, Takeda A, Chapman EJ, Sullivan CM, Fahlgren N, Brempelis KJ, Carrington JC (2010) Arabidopsis RNA-dependent RNA polymerases and dicer-like proteins in antiviral defense and small interfering RNA biogenesis during turnip mosaic virus infection. Plant Cell 22: 481–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Xu Y, Liu H, Mao Z, Zhang C, Ma Y, Zhang Q, Meng Z, Chong K (2013) The interaction between OsMADS57 and OsTB1 modulates rice tillering via DWARF14. Nat Commun 4: 1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey JJ, Lewsey MG, Patel K, Westwood J, Heimstädt S, Carr JP, Baulcombe DC (2011) An antiviral defense role of AGO2 in plants. PLoS ONE 6: e14639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Dong Z, Jia Z, Wang J, Wang G (2010) Isolation, expression and functional analysis of a putative RNA-dependent RNA polymerase gene from maize (Zea mays L.). Mol Biol Rep 37: 865–874 [DOI] [PubMed] [Google Scholar]
- He XF, Fang YY, Feng L, Guo HS (2008) Characterization of conserved and novel microRNAs and their targets, including a TuMV-induced TIR-NBS-LRR class R gene-derived novel miRNA in Brassica. FEBS Lett 582: 2445–2452 [DOI] [PubMed] [Google Scholar]
- Huber DM, Haneklaus S (2007) Managing nutrition to control plant Disease. Landbauforschung Völkenrode 57: 313–322 [Google Scholar]
- Huo Y, Liu W, Zhang F, Chen X, Li L, Liu Q, Zhou Y, Wei T, Fang R, Wang X (2014) Transovarial transmission of a plant virus is mediated by vitellogenin of its insect vector. PLoS Pathog 10: e1003949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Kitagawa M, Ihashi N, Yabe K, Kimbara J, Yasuda J, Ito H, Inakuma T, Hiroi S, Kasumi T (2008) DNA-binding specificity, transcriptional activation potential, and the rin mutation effect for the tomato fruit-ripening regulator RIN. Plant J 55: 212–223 [DOI] [PubMed] [Google Scholar]
- Katiyar-Agarwal S, Jin H (2010) Role of small RNAs in host-microbe interactions. Annu Rev Phytopathol 48: 225–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang H, Padmanabhan C, Li F, Kamei A, Bhaskar PB, Ouyang S, Jiang J, Buell CR, Baker B (2009) Identification of miniature inverted-repeat transposable elements (MITEs) and biogenesis of their siRNAs in the Solanaceae: new functional implications for MITEs. Genome Res 19: 42–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Pignatta D, Bendix C, Brunkard JO, Cohn MM, Tung J, Sun H, Kumar P, Baker B (2012) MicroRNA regulation of plant innate immune receptors. Proc Natl Acad Sci USA 109: 1790–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Lu YG, Shi Y, Wu L, Xu YJ, Huang F, Guo XY, Zhang Y, Fan J, Zhao JQ, et al. (2014) Multiple rice microRNAs are involved in immunity against the blast fungus Magnaporthe oryzae. Plant Physiol 164: 1077–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YF, Zheng Y, Addo-Quaye C, Zhang L, Saini A, Jagadeeswaran G, Axtell MJ, Zhang W, Sunkar R (2010) Transcriptome-wide identification of microRNA targets in rice. Plant J 62: 742–759 [DOI] [PubMed] [Google Scholar]
- Liu G, Ji Y, Bhuiyan NH, Pilot G, Selvaraj G, Zou J, Wei Y (2010) Amino acid homeostasis modulates salicylic acid-associated redox status and defense responses in Arabidopsis. Plant Cell 22: 3845–3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ZZ, Wang JL, Huang X, Xu WH, Liu ZM, Fang RX (2003) The promoter of a rice glycine-rich protein gene, Osgrp-2, confers vascular-specific expression in transgenic plants. Planta 216: 824–833 [DOI] [PubMed] [Google Scholar]
- Lu C, Jeong DH, Kulkarni K, Pillay M, Nobuta K, German R, Thatcher SR, Maher C, Zhang L, Ware D, et al. (2008) Genome-wide analysis for discovery of rice microRNAs reveals natural antisense microRNAs (nat-miRNAs). Proc Natl Acad Sci USA 105: 4951–4956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Yu H, Xiong G, Wang J, Jiao Y, Liu G, Jing Y, Meng X, Hu X, Qian Q, et al. (2013) Genome-wide binding analysis of the transcription activator IDEAL PLANT ARCHITECTURE1 reveals a complex network regulating rice plant architecture. Plant Cell 25: 3743–3759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez de Alba AE, Elvira-Matelot E, Vaucheret H (2013) Gene silencing in plants: a diversity of pathways. Biochim Biophys Acta 1829: 1300–1308 [DOI] [PubMed] [Google Scholar]
- Pumplin N, Voinnet O (2013) RNA silencing suppression by plant pathogens: defence, counter-defence and counter-counter-defence. Nat Rev Microbiol 11: 745–760 [DOI] [PubMed] [Google Scholar]
- Qi X, Bao FS, Xie Z (2009) Small RNA deep sequencing reveals role for Arabidopsis thaliana RNA-dependent RNA polymerases in viral siRNA biogenesis. PLoS ONE 4: e4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu F, Ye X, Morris TJ (2008) Arabidopsis DRB4, AGO1, AGO7, and RDR6 participate in a DCL4-initiated antiviral RNA silencing pathway negatively regulated by DCL1. Proc Natl Acad Sci USA 105: 14732–14737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Wang M, Meyerowitz EM (1996) DNA-binding properties of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA and AGAMOUS. Nucleic Acids Res 24: 3134–3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Ferrer V, Voinnet O (2009) Roles of plant small RNAs in biotic stress responses. Annu Rev Plant Biol 60: 485–510 [DOI] [PubMed] [Google Scholar]
- Saleh A, Alvarez-Venegas R, Avramova Z (2008) An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat Protoc 3: 1018–1025 [DOI] [PubMed] [Google Scholar]
- Satoh K, Kondoh H, Sasaya T, Shimizu T, Choi IR, Omura T, Kikuchi S (2010) Selective modification of rice (Oryza sativa) gene expression by rice stripe virus infection. J Gen Virol 91: 294–305 [DOI] [PubMed] [Google Scholar]
- Shivaprasad PV, Chen HM, Patel K, Bond DM, Santos BA, Baulcombe DC (2012) A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell 24: 859–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spann TM, Schumann AW (2010) Mineral nutrition contributes to plant disease and pest resistance. University of Florida IFAS Extension HS1181 [Google Scholar]
- Sunkar R, Girke T, Jain PK, Zhu JK (2005) Cloning and characterization of microRNAs from rice. Plant Cell 17: 1397–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Várallyay E, Válóczi A, Agyi A, Burgyán J, Havelda Z (2010) Plant virus-mediated induction of miR168 is associated with repression of ARGONAUTE1 accumulation. EMBO J 29: 3507–3519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waadt R, Schmidt LK, Lohse M, Hashimoto K, Bock R, Kudla J (2008) Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J 56: 505–516 [DOI] [PubMed] [Google Scholar]
- Wang N, Zhang D, Wang Z, Xun H, Ma J, Wang H, Huang W, Liu Y, Lin X, Li N, et al. (2014) Mutation of the RDR1 gene caused genome-wide changes in gene expression, regional variation in small RNA clusters and localized alteration in DNA methylation in rice. BMC Plant Biol 14: 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XB, Wu Q, Ito T, Cillo F, Li WX, Chen X, Yu JL, Ding SW (2010) RNAi-mediated viral immunity requires amplification of virus-derived siRNAs in Arabidopsis thaliana. Proc Natl Acad Sci USA 107: 484–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiberg A, Wang M, Bellinger M, Jin H (2014) Small RNAs: a new paradigm in plant-microbe interactions. Annu Rev Phytopathol 52: 495–516 [DOI] [PubMed] [Google Scholar]
- Wu L, Zhang Q, Zhou H, Ni F, Wu X, Qi Y (2009) Rice microRNA effector complexes and targets. Plant Cell 21: 3421–3435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Wang H, Hamera S, Chen X, Fang R (2014) miR444a has multiple functions in the rice nitrate-signaling pathway. Plant J 78: 44–55 [DOI] [PubMed] [Google Scholar]
- Yan Y, Zhang Y, Yang K, Sun Z, Fu Y, Chen X, Fang R (2011) Small RNAs from MITE-derived stem-loop precursors regulate abscisic acid signaling and abiotic stress responses in rice. Plant J 65: 820–828 [DOI] [PubMed] [Google Scholar]
- Yang SJ, Carter SA, Cole AB, Cheng NH, Nelson RS (2004) A natural variant of a host RNA-dependent RNA polymerase is associated with increased susceptibility to viruses by Nicotiana benthamiana. Proc Natl Acad Sci USA 101: 6297–6302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yuan YR, Pei Y, Lin SS, Tuschl T, Patel DJ, Chua NH (2006) Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev 20: 3255–3268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Su J, Duan S, Ao Y, Dai J, Liu J, Wang P, Li Y, Liu B, Feng D, et al. (2011) A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 7: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.