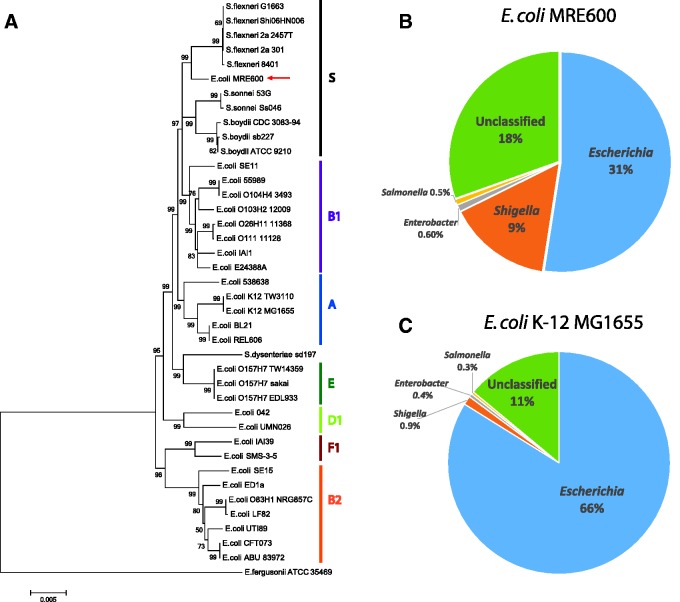
Fig. 3.—
Phylogenetic and taxonomic analysis of E. coli MRE600. (A) Phylogenetic tree showing the relationship of E. coli MRE600 with various strains of E. coli and Shigella based on multilocus sequence typing using seven housekeeping genes (adk, fumC, gyrB, icd, mdh, purA, and recA) (Wirth et al. 2006). The evolutionary history was inferred using the Neighbor-Joining method (Saitou and Nei 1987). Escherichia coli phylogenetic groups (A, B1, B2, D1, E, F1) as well as a Shigella group (S) are indicated. Escherichia fergusonii ATCC 35469 was used as the outgroup. The optimal tree with the sum of branch length = 0.17149911 is shown. The confidence probability (multiplied by 100) that the interior branch length is greater than 0 was estimated using the bootstrap test (1,000 replicates) and is shown next to the branches (Dopazo 1994; Rzhetsky and Nei 1992). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Maximum Composite Likelihood method (Tamura et al. 2004) and are in the units of the number of base substitutions per site. The analysis involved 40 sequences (9,015 positions each), and positions containing gaps and missing data were eliminated. All analyses were conducted in MEGA6 (Tamura et al. 2013). (B, C) Sequence classification of MRE600 and E. coli K12 using Kraken (Wood and Salzberg 2014). The filtered subreads used for this K12 analysis were from a 125× coverage assembly of E. coli K-12 MG1655, which we generated in the same manner as that for MRE600. Pie charts highlight the top four genera associated with the indicated strains. (B) MRE600 shows greater similarity to Escherichia (31% matches) than Shigella (9% matches). (C) Escherichia coli K12 also showed the greatest similarity to Escherichia (66%), but its match with Shigella was much lower (0.9%).