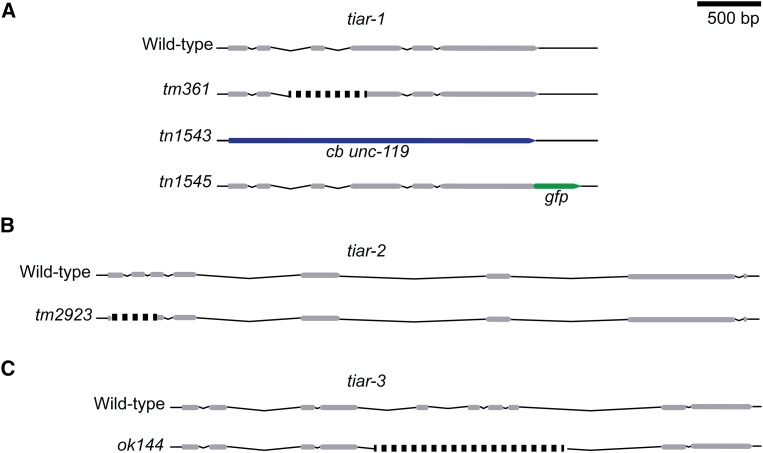
Figure 2.
Alleles used to study the function of tiar genes in C. elegans. (A) The tiar-1 wild-type allele comprises six exons (gray) and five introns (thin peaks). The tiar-1(tm361) allele is an out-of-frame deletion of 581 bp (dotted line). We generated a deletion and a GFP fusion allele using CRISPR-Cas9 genome editing. The strain tiar-1(tn1543) results from a deletion of the tiar-1 open reading frame. In this strain, tiar-1 sequences were replaced with the C. briggsae unc-119 gene as a positive selection marker (blue). The strain tiar-1(tn1545) has an in-frame insertion of the tag S::TEV::GFP at the TIAR-1 C- terminus (green). (B) The tiar-2 wild-type allele comprises eight exons and eight introns. The tiar-2(tm2923) is a deletion of 336 bp (dotted line). (C) The tiar-3 wild-type allele comprises 10 exons and nine introns. The tiar-3(ok144) is a deletion of 1531 bp (dotted line).