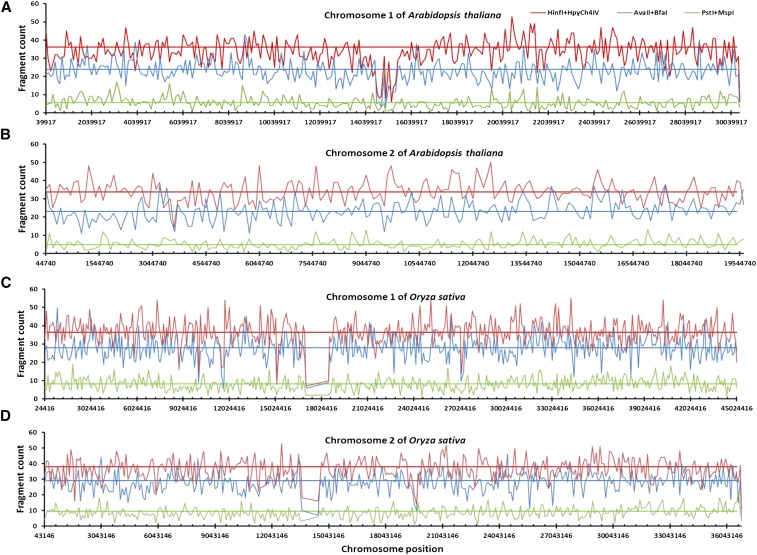
Figure 2.
Fragment distributions detected in silico on selected chromosomes of Arabidopsis and rice. Distributions of DNA fragments generated by in silico digestions with three restriction enzyme combinations on two chromosomes of Arabidopsis thaliana (A, B) and Oryza sativa (C, D). The number of DNA fragments and the average enzyme-cutting position based on a 100 kb sliding-window of a given chromosome are calculated and shown with a colored line for each enzyme combination. The corresponding horizontal linear line represents the average fragment count for an enzyme combination on the chromosome. More digestions were found for HinfI + HpyCh4IV than the other two enzyme combinations.