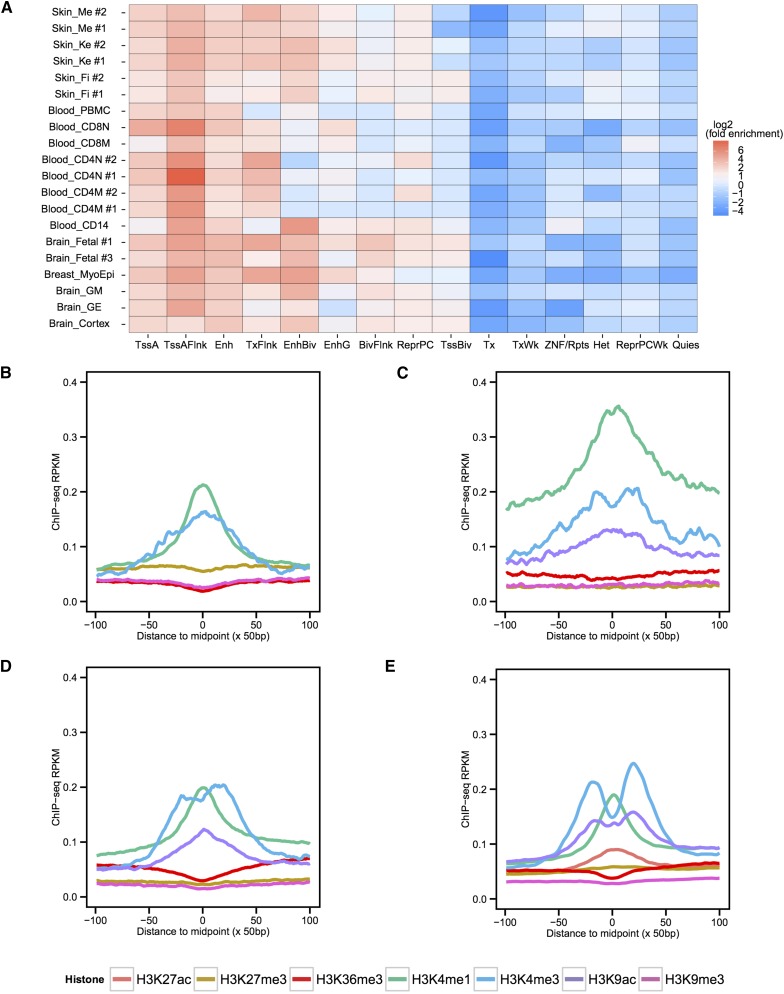
Figure 4.
Variably methylated regions (VMRs) enrich for enhancer chromatin states and relevant histone modifications. (A) Enrichment of 15-state ChromHMM annotations in cell type-specific hypomethylated VMRs. Log2 transformed fold enrichment values for each ChromHMM state in each cell type were calculated and plotted. Replicate ChromHMM states were used for skin melanocytes, fibroblasts, keratinocytes, CD4 naïve cells, CD4 memory cells, and fetal brain, with replicate number indicated after cell type abbreviation detailed in Table S1. TssA, Active TSS; TssAFlnk, Flanking active TSS; Enh, Enhancers; TxFlnk, Transcribed at gene 5′ and 3′; EnhBiv, Bivalent enhancer; EnhG, Genic enhancers; BivFlnk, Flanking bivalent TSS/Enh; ReprPC, Repressed Polycomb; TssBiv, Bivalent/poised TSS; Tx, Strong transcription; TxWk, Weak transcription; ZNF/Rpts, ZNF genes + repeats; Het, Heterochromatin; ReprPCWk, Weak repressed Polycomb; Quies, Quiescent. ChIP-seq signal density RPKM values over a 10 kb region centered on hypomethylated VMRs for (B) cortex derived neurosphere cells, (C) CD8 naïve T cells, (D) breast myoepithelial cells, and (E) skin keratinocytes. RPKM values at 50 bp resolution were calculated and plotted.