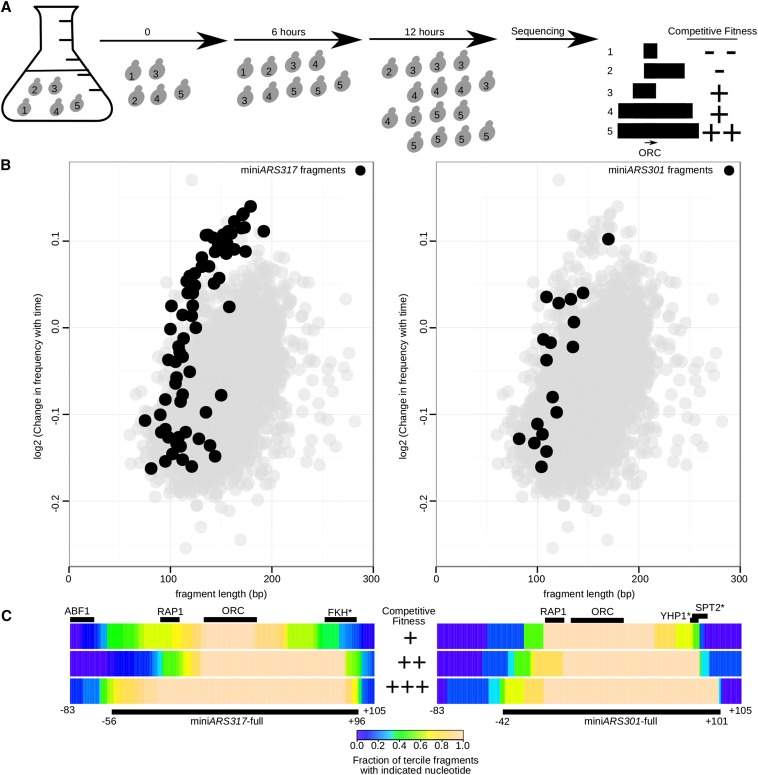
Figure 1.
The ARSs associated with the HMR-E and HML-E transcriptional silencers generated many of the most competitive miniARS fragments in the miniARS library. (A) A pool of transformed yeast was grown competitively under conditions that selected for the plasmid. Deep sequencing measured the frequency of the miniARS fragments within the population throughout the course of the experiment. In this simple example, miniARS fragment #5 is enriched during the course of the experiment and is assigned a positive competitive fitness value. In contrast, miniARS fragment #2 is depleted during the course of the experiment and is assigned a negative competitive fitness value. If these fragments were from a single ARS (autonomously replicating sequence), ranking them based on their competitive fitness values would help indicate the minimal chromosomal region necessary for maximizing the function of this ARS. In addition, competitive fitness between origins can be compared to allow identification of the most competitive ARSs in the population. (B) The slope of the log2 ratios of the frequency of each miniARS fragment in the yeast cell population through the time course was used to measure each fragment’s competitive fitness (CF). This value is plotted against the length of the fragment in bp (x-axis). A gray circle designates each fragment that was assessed in this miniARS experiment. In the left panel, the HMR-E silencer-associated fragments (miniARS317) present in the experiment are filled in black, while in the right panel the HML-E silencer-associated fragments (miniARS301) are filled in black. (C) All of the miniARS317-fragments (n = 70, left panel) and all of the miniARS301-fragments (n = 18, right panel) were ranked based on their CF values and then divided into three distinct bins based on this value. These bins were then ranked based on their average competitive fitness (CF) values from lowest (+) to highest CF (+++). To better visualize nucleotides that were enriched in the most competitive miniARS for each silencer, an arbitrary fragment was selected encompassing each silencer and numbered relative to the first nucleotide of the T-rich strand of the ORC (origin recognition complex) binding site that was given the value “0.” Each bin was color coded as indicated to visualize the fraction of fragments within that bin that contained a given nucleotide. Thus, the most competitive bin of miniARS317 fragments indicated that greater than 80% of the fragments contained intact RAP1 (Rap1 protein binding site), ORC (origin recognition complex), and FKH (forkhead) sites. The most competitive miniARS silencers (miniARS317-full and miniARS301-full) used for deep mutational scanning in Figure 2 are indicated below the most competitive tercile with a thick black line. Their numbering is relative to the ORC binding site, as above. The FKH*, YHP1* (Yeast Homeo-Protein), and SPT2 (SuPpressor of Ty’s)* sites are starred to indicate that their identification is based solely on a strong motif match. In contrast, the silencer ABF1 (ARS-Binding Factor 1), RAP1, and ORC sites have been verified as such by numerous genetic and biochemical experiments.