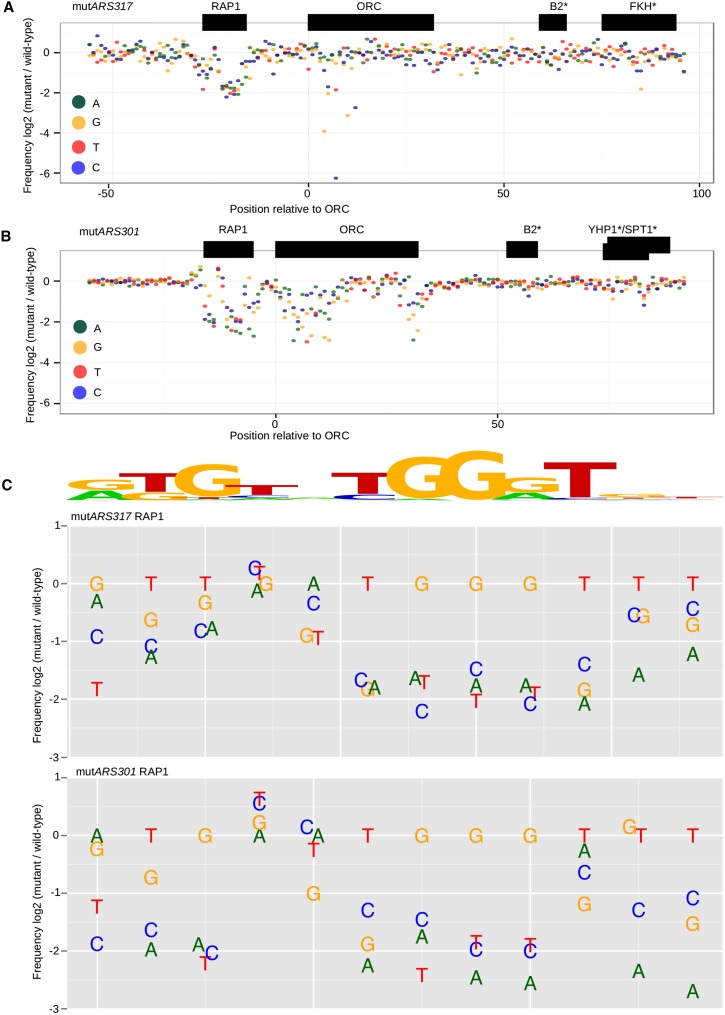
Figure 2.
Deep mutational scanning of the HMR-E and HML-E silencer miniARSs-defined nucleotides required for their competitive fitness. (A) The data for a library of miniARS317-mut fragments was graphed to indicate the individual nucleotide substitutions that affected the competitive fitness of miniARS317-full. (B) As in 2A except data are from a miniARS301-mut experiment. (C) Detailed analyses of the mutational profile of the RAP1 (Rap1 protein binding site) sites. A consensus RAP1 site derived from genome-wide chromatin immunoprecipitation data are shown above the profiles. FKH, forkhead; ORC, origin recognition complex; YHP1, Yeast Homeo-Protein; SPT2, SuPpressor of Ty’s.