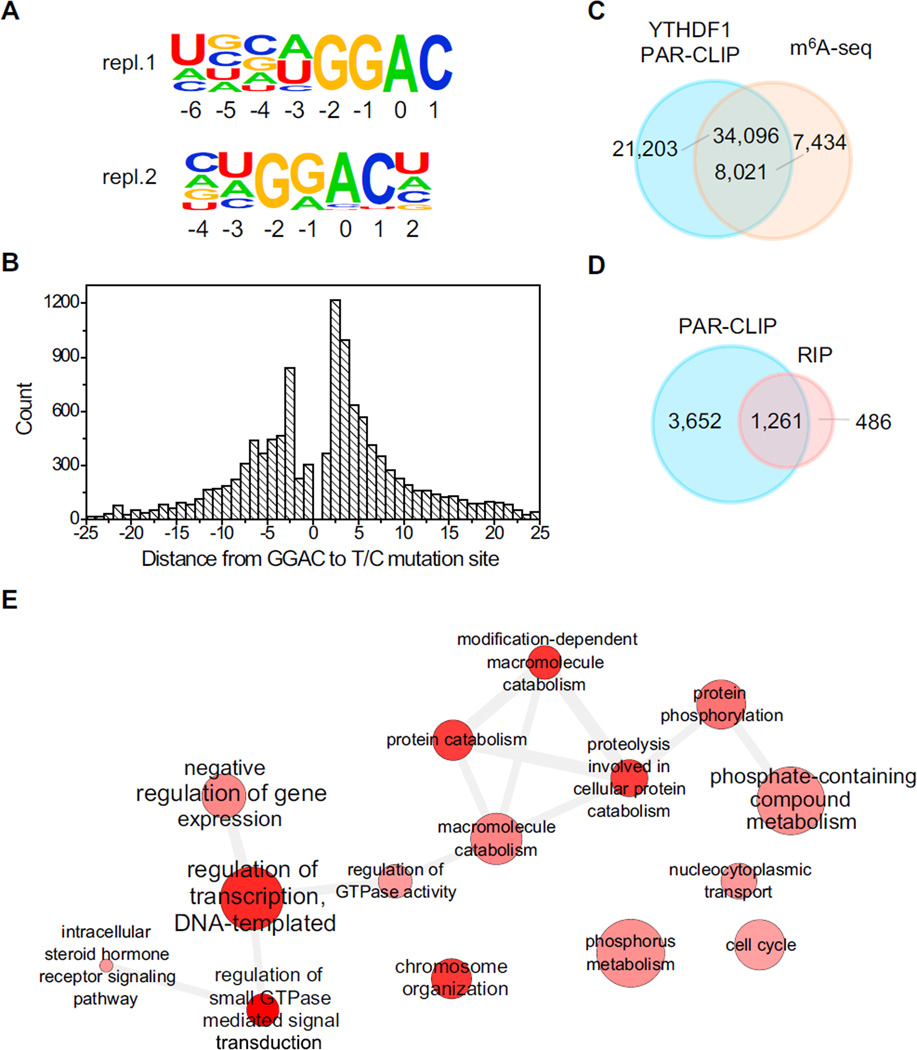
Figure 1. Transcriptome-wide Identification of YTHDF1 mRNA Targets.
(A) YTHDF1-binding motifs identified by HOMER from PAR-CLIP peaks of two biological replicates. Motif length was restricted to 6–8 nucleotides. The motif with the lowest p value of replicate 1 (repl.1) was found in 53.8% of 24,753 sites (p = 1 × 10−703), and that of replicate 2 (repl.2) was found in 54.5% of 58,549 sites (p = 1 × 10−1093).
(B) The distribution of the distance from GGAC to T-to-C mutation sites. The optimum crosslinking sites are at the −3, +2, and +3 positions.
(C) Overlap of YTHDF1 PAR-CLIP peaks and m6A-seq peaks in HeLa cells.
(D) Overlap of target genes identified by PAR-CLIP and RIP-seq for YTHDF1.
(E) Gene Ontology analysis of YTHDF1 mRNA targets.
See also Figure S1 and Table S1.