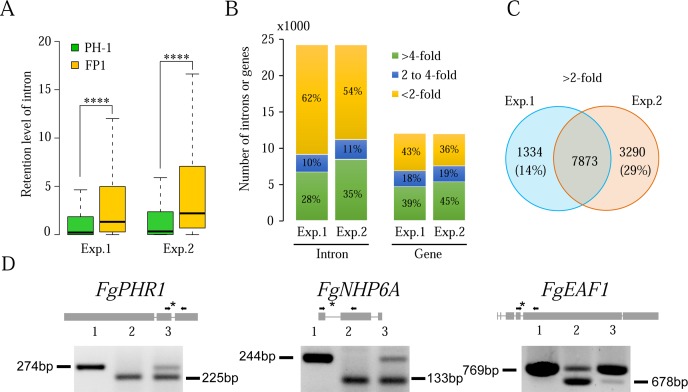
Fig 3. Effects of FgPRP4 deletion on intron splicing.
(A). Box-plot comparison of intron retention levels between the wild type (PH-1) and Fgprp4 mutant (FP1) in replica experiments. The statistical significance for each comparison is analyzed by t-test (****, P<0.0001). (B). The percentage of introns and genes with the three marked intron retention levels in Fgprp4 compared to the wild type. (C). Introns that were increased in intron retention over 2-fold in Fgprp4 in both replica experiments. (D). Intron splicing defects in the labelled genes were verified by RT-PCR with primers flanking the introns with reduced splicing efficiency (marked with *) in the Fgprp4 mutant. Lanes 1–3 were PCR results with the genomic DNA, cDNA of PH-1, and cDNA of Fgprp4, respectively. The sizes of amplified bands are labelled on the side.