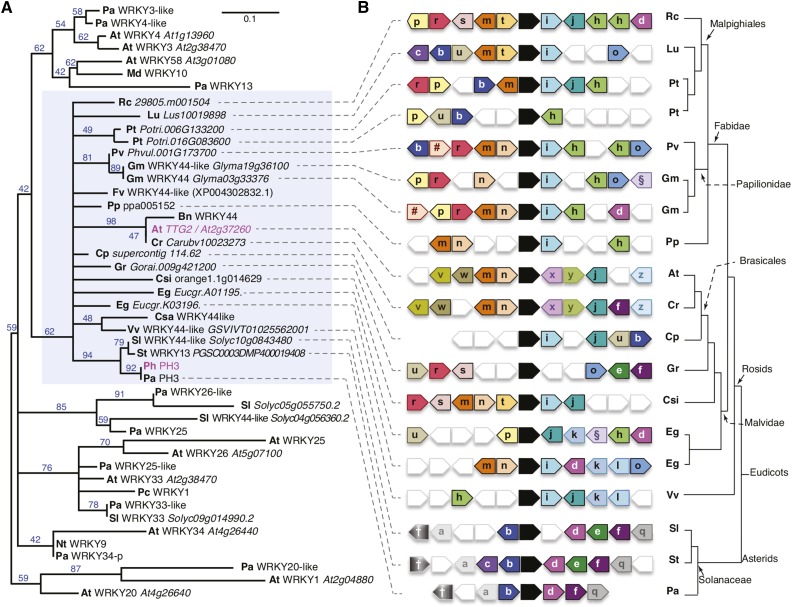
Figure 3.
PH3 Is a WRKY Protein and Homologous to Arabidopsis TTG2.
(A) Phylogenetic tree of a selection of type I WRKY proteins. Blue numbers on tree branches indicate percentage of bootstrap support (1500 replicates). The blue shaded rectangle indicates the clade containing TTG2/PH3 homologs. Prefixes denote the species of origin: Arabidopsis thaliana (At), Brassica napus (Bn), Capsella rubella (Cr), Carica papaya (Cp), Citrus sinensis (Csi), Cucumis sativum (Csa), Eucalyptus grandis (Eg), Fragaria vesca (Fv), Glycine max (Gc), Gossypium raimondii (Gr), Hordeum vulgare (Hv), Ipomea batatas (Ib), Linum usitatissimum (Lu), Malus domestica (Md), Nicotiana tabacum (Nt), Petroselinum crispum (Pc), Petunia axillaris (Pa), Petunia hybrida (Ph), Phaseolus vulgaris (Pv), Populus trichocarpa (Pt), Prunus persica (Pp), Ricinus communis (Rc), Solanum lycopersicum (Sl), Solanum tuberosum (St), and Vitis vinifera (Vv).
(B) Diagram showing relationships between genes immediately surrounding putative TTG2/PH3 homologs from distinct species inferred by Phytozome (Goodstein et al., 2012). Analysis of the P. axillaris region was done manually using unpublished sequence data from the petunia platform. The arrows denote genes and direction of transcription. Black arrows indicate PH3/TTG2 homologs. Similarity among flanking genes is indicated by similar colors and symbols/letters. Relationships of the different (groups of) species of origin are indicated on the right.