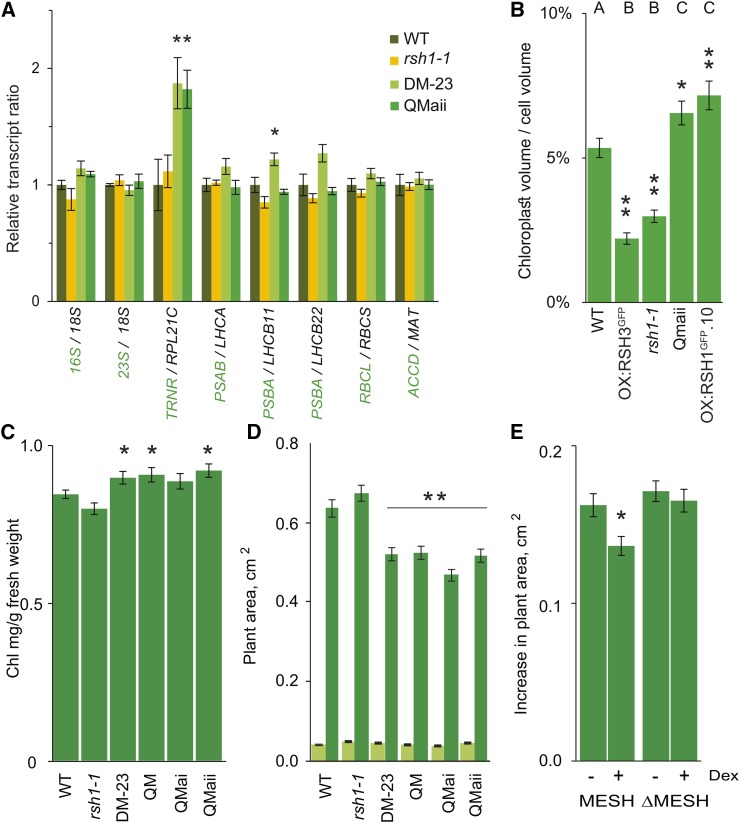
Figure 6.
RSH Enzymes Are Required for Regulating Chloroplast Function, Volume, and Plant Growth.
(A) Ratios of chloroplast (green) to nucleus-encoded (black) transcripts in different RSH mutants. qRT-PCR was performed on cDNA extracted from seedlings grown on plates for 12 DAS. Data are presented as means ± se for five independent biological replicates. Significance was calculated using ANOVA with post-hoc Dunnett tests versus the wild-type controls.
(B) Total chloroplast volume per cell was calculated in protoplasts isolated from fully expanded leaves of plants grown on soil at 35 DAS. Representative protoplast images are shown in Supplemental Figure 9B. Statistical significance was calculated using Kruskal-Wallis with the Dunn test post hoc, and the resulting groups are indicated above each bar. Thirty protoplasts were analyzed for OX:RSH3-GFP.1 (OX:RSH3GFP) and 47 to 59 for the other lines. Similar results were also obtained using an independent approach on intact cells (Supplemental Figure 9C).
(C) Chlorophyll levels were measured in selected RSH mutants grown on soil at 24 DAS. DM-23 and the QMs have a higher chlorophyll content than the wild type; two-way Student’s t test, n = 8 plants.
(D) Plant surface area for wild-type and mutant seedlings grown on plates at 6 (light green) and 12 DAS (dark green) after stratification. Except for rsh1-1, which was larger (P < 0.0001), there were no significant differences between the mutants and the wild type at 6 DAS. Similar results were also obtained for plants grown in soil (Supplemental Figure 9D). Significance was calculated using ANOVA with post-hoc Dunnett tests versus the wild-type controls; n = 50 plants per line.
(E) At 8 DAS, MESH and ΔMESH plants were transferred onto medium containing DMSO (control) or 1 µM dexamethasone (induced) and the increase in plant area was measured 4 d after transfer. The two-way Student’s t test was used to compare noninduced and induced plants. n = 36 plants,*P < 0 .05 and **P < 0.01, error bars indicate se.