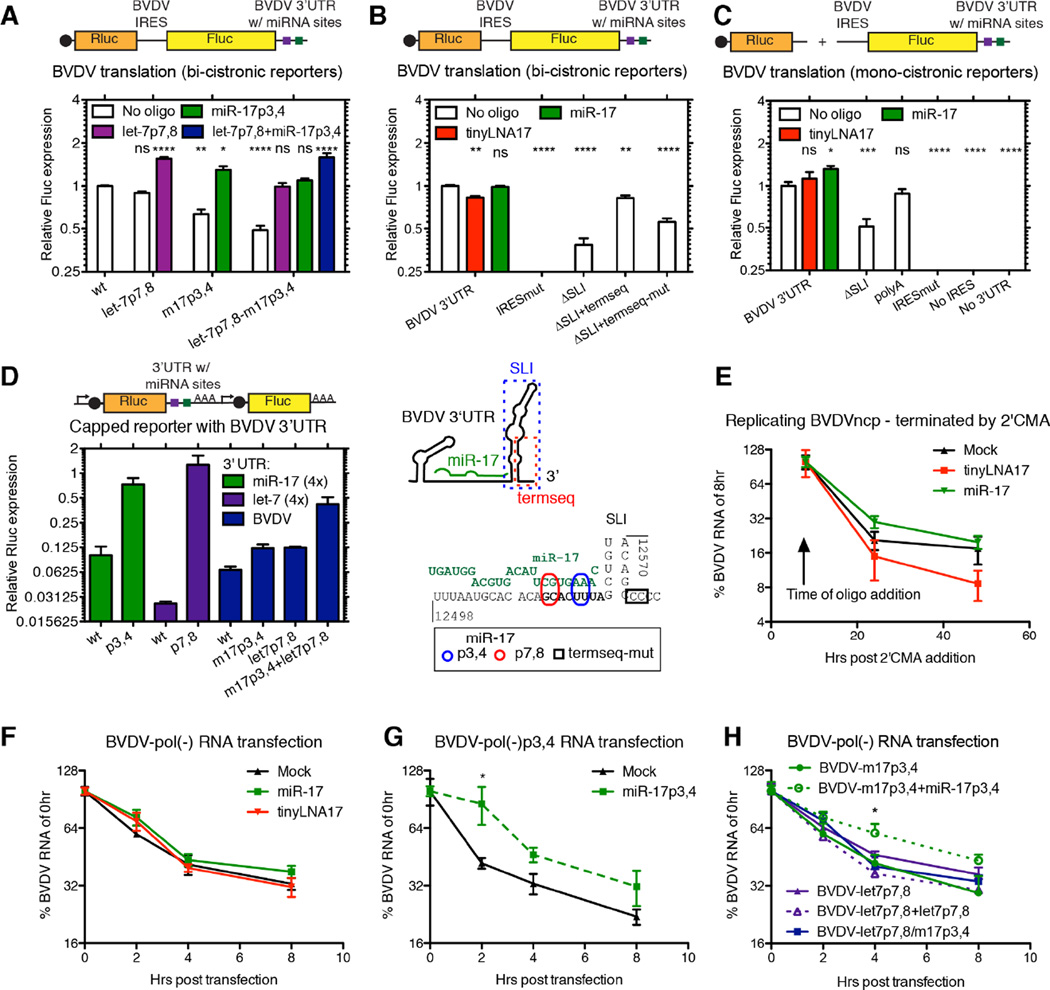
Figure 6.
Functional characterization of BVDV miRNA interactions. (A) BVDV driven translation was measured from bicistronic reporters after introduction of let-7 or miR-17 seed site mutations and after trans-complementation with miRNA mimics (50nM). (B) Translation as in (A) measured after treatment with tinyLNA-17 (0.3µM) or miR-17 (50nM) or after modifications to the 3’ UTR as indicated in the schematic below. (C) Translation as in (B), but measured from monocistronic reporters with the exact BVDV termini. (D) Translation measured from RNAs harboring the BVDV 3’ UTR and/or miRNA seed sites in the capped, poly(A)-tailed context of the psiCHECK-2 system. In the schematics, caps are black circles, miRNA sites are squares, promoters are angled arrows, and poly(A)-tails are AAA’s. (E–G) Effect of miRNAs on BVDV RNA stability. Decay of BVDV RNA was measured +/− tinyLNA-17 (0.3µM) or miR-17 (50nM) after blocking active BVDV replication using 2’CMA (E), or after transfection of replication-deficient pol(-)BVDV RNA (F). (G–H) RNA decay assays as in (F), but for seed site mutants +/− corresponding miRNA complementation. See also Figure S6.