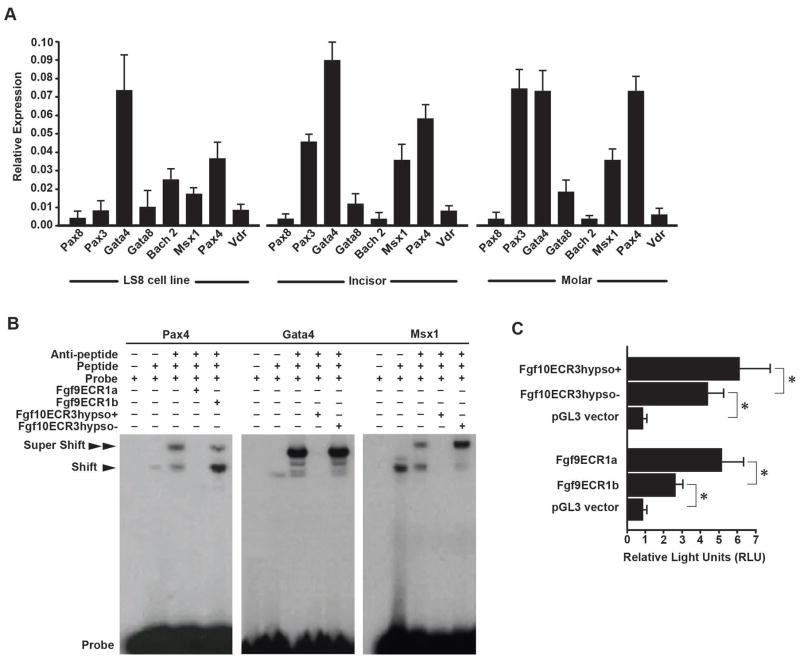
Figure 5.
Analysis of enhancer function of Fgf9ECR1 and Fgf10ECR3. (A) rtPCR expression data of transcription factors associated with the ECRs in LS8 ameloblast cells, adult mouse incisor cervical loops (incisor), and E16.5 mouse molars. (B) EMSA shows that Fgf9ECR1a competes with the control probe for interaction with PAX4, and Fgf10ECR3hypso+ competes with the control probe for interaction with MSX1, and GATA4, while Fgf9ECR1b and Fgf10ECR3hypso− do not. (C) Luciferase assay conducted in LS8 cells shows stronger enhancer function in Fgf9ECR1a and Fgf10ECR3hypso+ than Fgf9ECR1b and Fgf10ECR3hypso− (*p<0.001).