Figure 4. miR-101 reversed the hypomethylation status of the PRDM16 promoter.
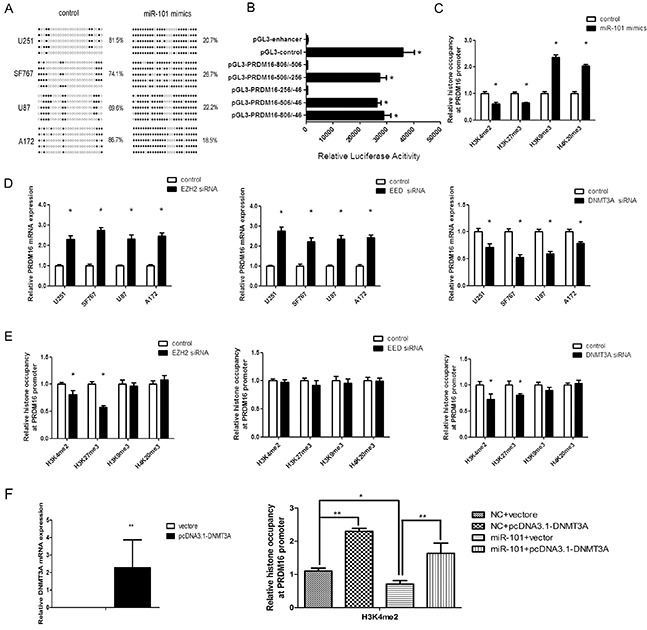
A. PRDM16 methylation levels were increased by miR-101 in U251 cells. The unmethylated and methylated CpG sites are indicated by opened and closed circles, respectively. Each row indicates the sequencing result of one clone of the bisulfite-PCR product. The number of methylated CpGs was divided by the total number of true CpGs analyzed and is given as a percentage to the right of each BSP result. B. Analysis of promoter activity of the PRDM16 core promoter constructs via luciferase reporter assays. The construct containing the sequence spanning the region from −506 to −256 was sufficient to mediate maximal promoter activity. The core promoter ranged from −506 to −256. PGL3-control is the positive control, and pGL3-enhancer is the negative control. An independent samples t-test was used. *P<0.05. C. The histones occupancy of the PRDM16 promoter was affected by miR-101. A ChIP assay was used to detect the H3K4me2, H3K27me3, H3K9me3 and H4K20me3 occupancy the PRDM16 core promoter. U251 cells were transfected with miR-101 or a scrambled control. An independent samples t-test was used. *P<0.05. D. PRDM16 expression was regulated by EZH2, EED and DNMT3A. Real-time PCR analysis was performed 48 h after transfection with EZH2 siRNA, EED siRNA, DNMT3A siRNA or a scrambled control. An independent samples t-test was used. *P<0.05. E. The histones occupancy of the PRDM16 promoter was affected by EZH2 siRNA, EED siRNA and DNMT3A siRNA. A ChIP assay was performed to detect the H3K4me2, H3K27me3, H3K9me3 and H4K20me3 occupancy of the PRDM16 core promoter. U251 cells transfected with EZH2 siRNA, EED siRNA and DNMT3A siRNA were analyzed. An independent samples t-test was used. *P<0.05. F. Left: Real-time PCR was used to detect DNMT3A expression after transfection with a vector or pcDNA3.1-DNMT3A. Right: a ChIP assay was used to detect the H3K4me2 occupancy of the PRDM16 core promoter. U251 cells transfected with miR-101 and pcDNA3.1-DNMT3A were analyzed.
