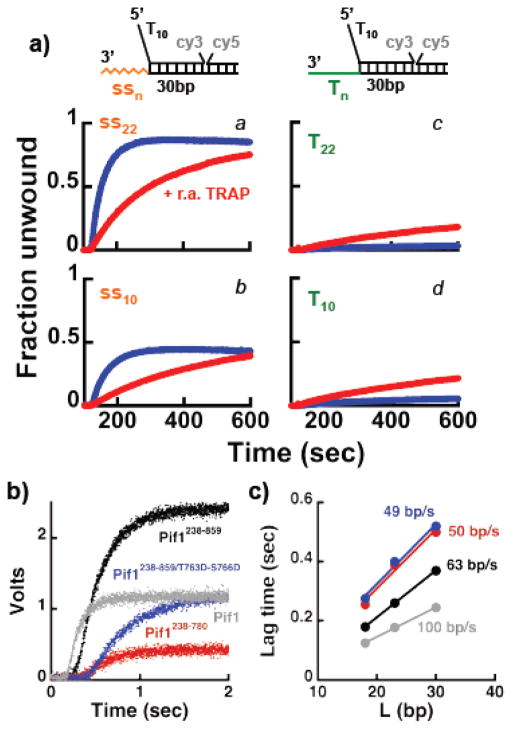
Figure 6. Deletion of the C-terminus of Pif1 affects the unwinding activity of the monomer. a).
FRET-based unwinding experiments under multiple turnover conditions using 15 nM Pif1238–780 and 20 nM of the indicated substrates, starting the reaction by addition of either 0.5 mM ATP (blue) or 0.5 mM ATP and 3.5x of the r.a. TRAP. b) Stopped-flow experiment comparing the unwinding activity of a monomer of different Pif1 constructs (75 nM) using a substrate (100 nM) with a 23 bp dsDNA, a 5′-dT10 and a 22 nt 3′-tail of mixed sequence composition. c) Lag times as a function of dsDNA length obtained at the higher concentrations used in b). The calculated rate of unwinding of the different Pif1 constructs is shown, with the colors matching the ones in b).