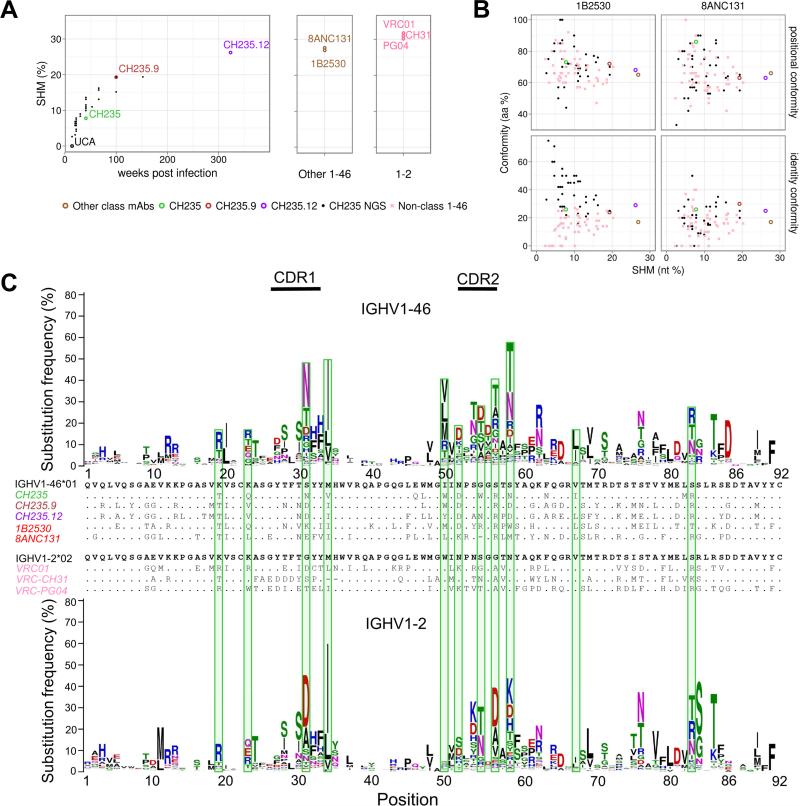
Figure 3. Sequence Evolution of CH235 Lineage: SHM, Timing, and Conformity of CH235-Lineage Development from UCA to Antibody with 90% Breadth.
(A) Heavy chain SHM over time for the CH235 lineage (left panel). SHM levels of other VH1-46-derived CD4bs mAbs and selected VH1-2-derived VRC01-class mAbs are shown (middle and right panels, respectively); the time since infection is unknown for these mAbs. (B) Maturation conformity vs overall heavy chain SHM. Positional conformity (top row) is defined as the number of aa positions differing from the germline sequence in both the conforming and reference sequences, divided by the total number of aa changes in the conforming antibody. Identity conformity (bottom row) is defined as the number of such positions which are additionally mutated to the same residue, divided by the total number of mutations in the conforming antibody. Conformity to 1B2530 (left) and to 8ANC131 (right) is shown for both position and identity. (C) VH-gene mutability accounts for the majority of positional conformity of CH235 lineage. The mutability of the VH-gene for VH1-46 (top) and VH1-2 (bottom) is shown. Sequence logos are shown at each position; the height of each logo corresponds to the percent of mutated reads. Green bars are shown for SHM in antibody CH235, which are altered in over a quarter of VH1-46-derived antibodies. See also Figure S3 and Table S4.