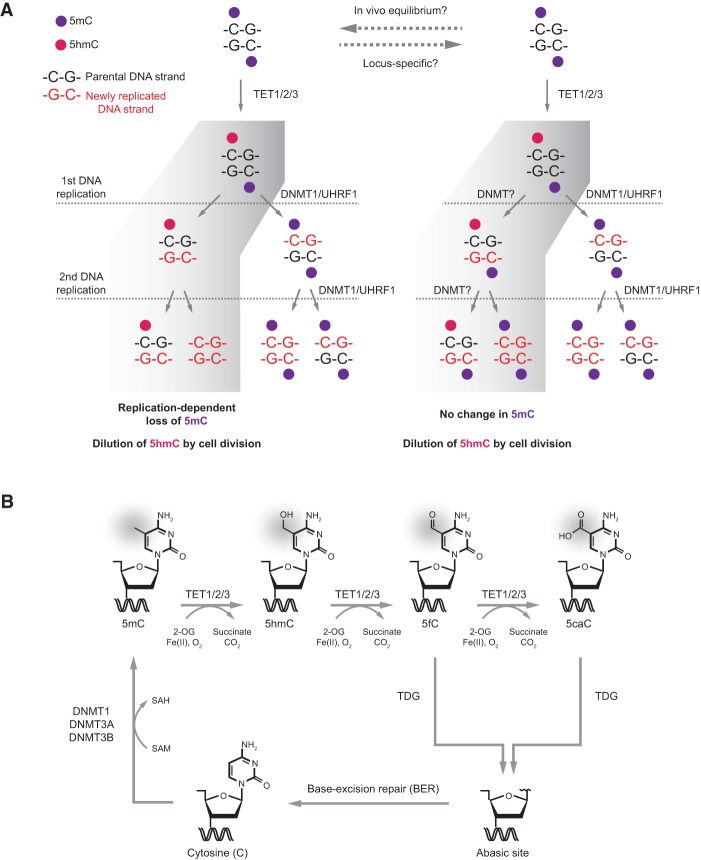
Figure 2.
Pathways of DNA demethylation mediated by TET enzymes. (A) Model of passive replication-dependent DNA demethylation. The diagrams illustrate the two opposite fates of a hemihydroxymethylated CpG dinucleotide through two rounds of DNA replication. The left panel shows the replication-dependent DNA demethylation, in which DNA methylation is lost on the DNA strand opposite to 5hmC. The right panel shows the reverse outcome, in which maintenance of DNA methylation is achieved by DNMT1 in complex with UHRF1/2 or DNMT3A/B. The relative extent of these two opposing outcomes of 5hmC deposition in vivo remains to be determined and is likely influenced by global as well as locus-specific factors. In both cases, newly replicated DNA dilutes 5hmC during cell division. (B) Model of active DNA demethylation by a TET/TDG (thymine–DNA–glycosylase)/BER (base excision repair)-dependent pathway. A cytosine base can be methylated by the DNA methylation machinery (DNMT1 or DNMT3A/B) to form 5mC, which in turn can be iteratively oxidized by TET enzymes to produce 5hmC, 5fC, and 5caC. TDG then recognizes 5fC and 5caC, and the oxidized cytosine base is excised. This yields an abasic site that is repaired by BER and results in restoration of the unmodified cytosine state. Additional pathways of active DNA demethylation have been suggested (for review, see Wu and Zhang 2014). (SAH) S-adenosyl-homocysteine; (SAM) S-adenosyl-methionine.