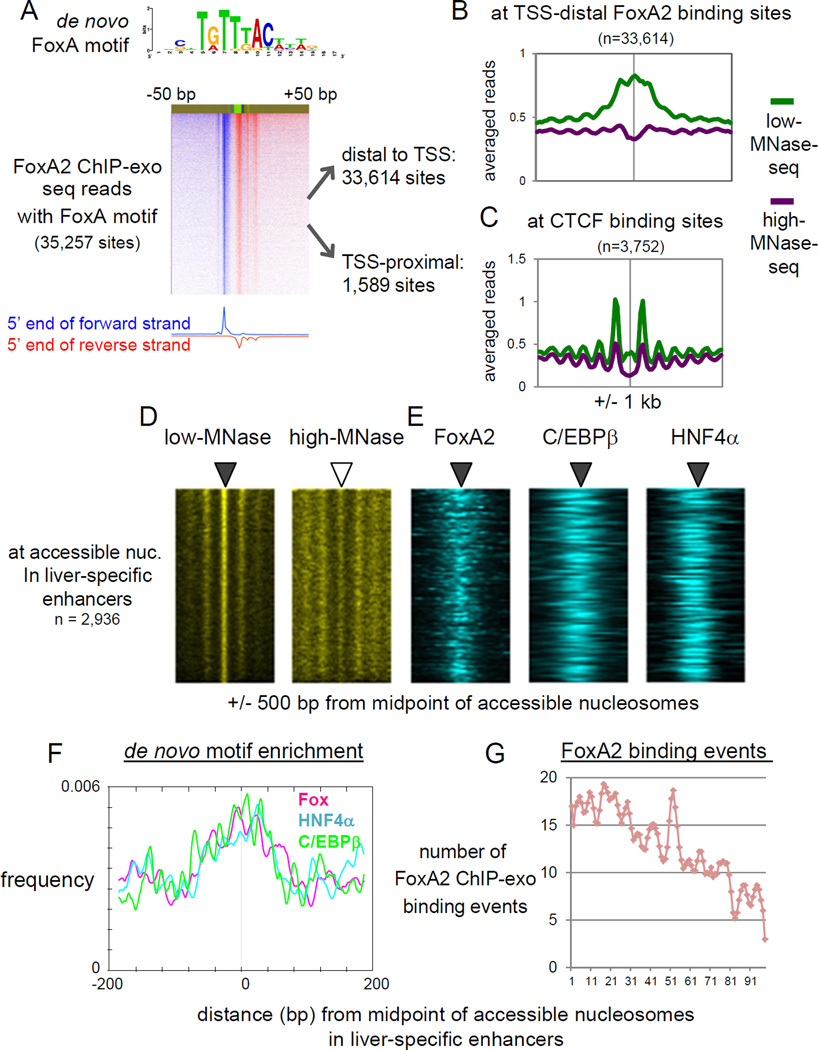
Figure 3. Pioneer factor FoxA2 and liver TFs are co-localized at accessible nucleosomes in liver-specific enhancers.
(A) FoxA2 ChIP-exo tag 5’ end distribution centered by de novo FoxA motif midpoint and sorted by FoxA2 occupancy level. (B) Averaged profiles of low- and high-MNase-seq at TSS-distal FoxA2 binding sites, and (C) at CTCF binding sites overlapping with H3K27ac peaks. (D. E) Heatmaps of MNase-seqs, and FoxA2 ChIP-exo, C/EBβ and HNF4α ChIP-seq at MNase-accessible nucleosomes in liver-specific enhancers, rank ordered by low-MNase-seq read density. (F) Profiles of de novo motifs enrichment relative to midpoint of accessible nucleosome in enhancers. (G) Histogram of FoxA2 ChIP-exo binding locations relative to midpoint of accessible nucleosomes. The midpoint approximating the position of the respective nucleosome dyad axis. (See also Figure S3)