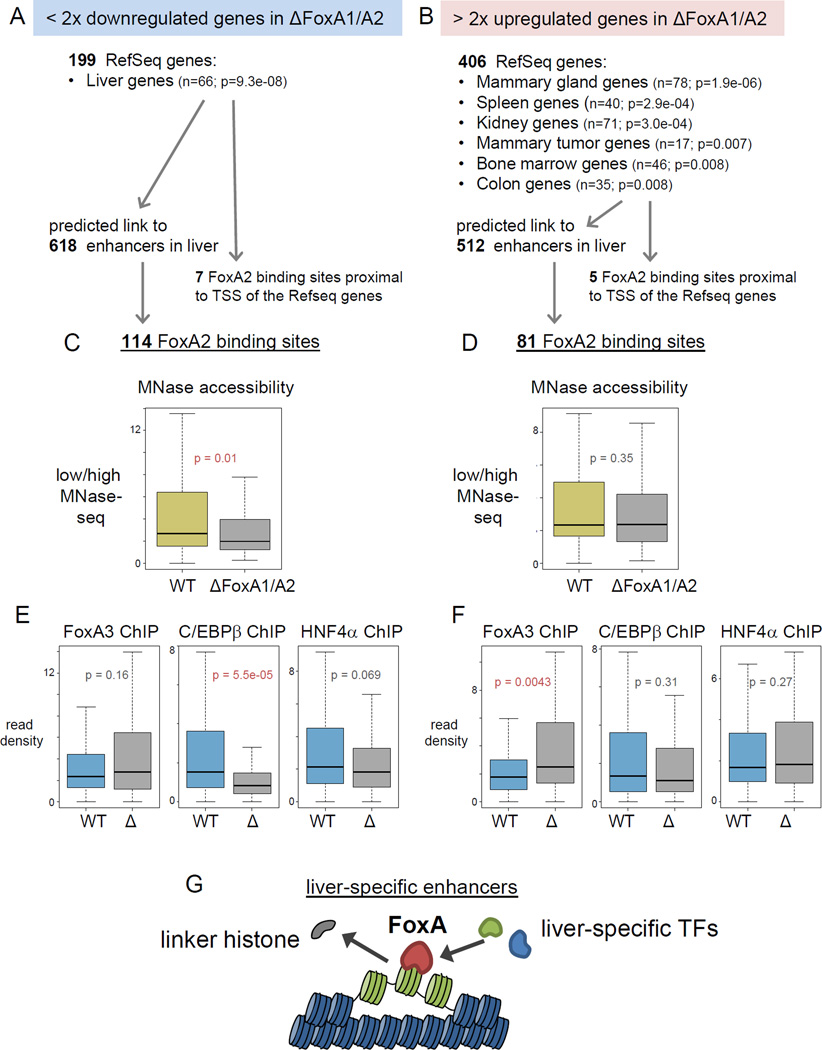
Figure 7. Maintenance of accessible nucleosomes by FoxA is required for liver-specific gene activation.
(A, B) Tissue Expression analysis for > two-fold upregulated or downregulated genes in the FoxA1/A2 deletion mutant (Li et al., 2011) (See also Table 3). Number of enhancers that potentially link to the target genes (Shen et al., 2012) and number of FoxA2 ChIP-exo sites in the enhancers. (C, D) Box and whiskers plot show MNase accessibility (low/high MNase-seq) in wild type and the FoxA1/A2 mutant at central 200 bp of downregulated genes related FoxA2 sites and (D) at upregulated genes related FoxA2 sites (See also Figure S7). (E) Box and whiskers plot show FoxA3, C/EBβ and HNF4α ChIP-seq in wild type (WT) and the FoxA1/A2 mutant (Δ) at central 200 bp of downregulated genes related FoxA2 sites and (F) at upregulated genes related FoxA2 sites. p-values by Wilcoxon rank-sum test. (G) Models of nucleosome configuration at liver-specific enhancers. FoxA2 binding is required to displace linker histones, keep nucleosome accessible, and help recruitment of other liver enriched TFs, which stimulate liver gene transcription.