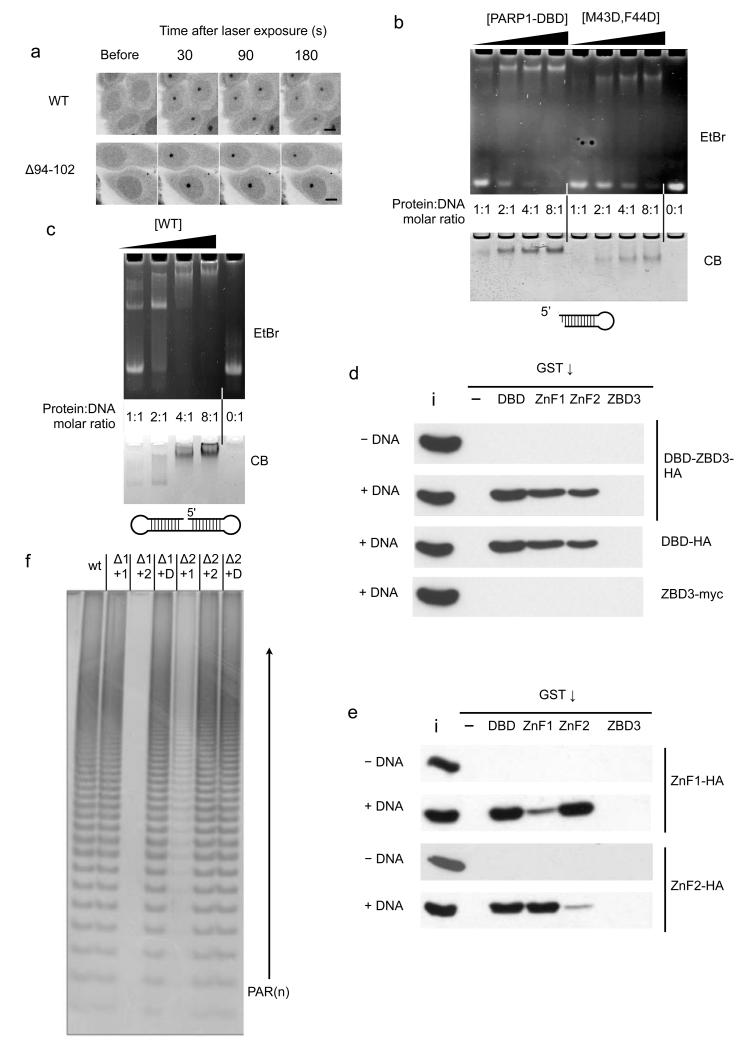
FIGURE 5. Intermolecular dimerisation of PARP1 Znf1 and ZnF2 domain.
a) A nine residue deletion in the linker connecting ZnF1 and ZnF2 has no effect on the ability of the PARP1 - DBD to localise to damage foci. Scale bar indicates 10 μm.
b) Wild-type PARP1-DBD (WT) forms a strong interaction with the DNA in an electrophoretic mobility assay (EMSA) that is only saturated at a 2:1 protein:DNA ratio. DNA binding of the M43D, F44D PARP1-DBD mutant is substantially impaired.
c) EMSA of a ‘dumbbell’ oligonucleotide shows some complex formation at 1:1 protein-DNA ratio, but a 2:1 ratio is required to achieve saturation.
d) Pull-down assay showing that the DBD and isolated DNA-binding zinc-finger domains, but not the third zinc-binding domain (ZBD3), are able to co-precipitate constructs incorporating the DBD in a DNA dependent manner. ‘i’ = 20% input, ‘-‘ = empty beads.
e) Pull-down assay showing that either of the isolated tagged DNA-binding zinc-finger domains can be efficiently co-precipitated in a DNA-dependent manner, by the GST-DBD fusion or the GST-fusion with the other zinc-finger domain, but not by the same zinc-finger domain or by GST-ZBD3. ‘i’ = 5% input, ‘-‘ = empty beads.
f) Catalytic activity of PARP1 constructs deleted for either ZnF1 (Δ1) or ZnF2 (Δ2) domain can be complemented by presentation of the deleted domain in trans (+1, +2, or D = +1 and 2), consistent with the requirement for both DNA-binding zinc-finger domains to form an intermolecular interaction for PARP1 activation at sites of DNA damage.