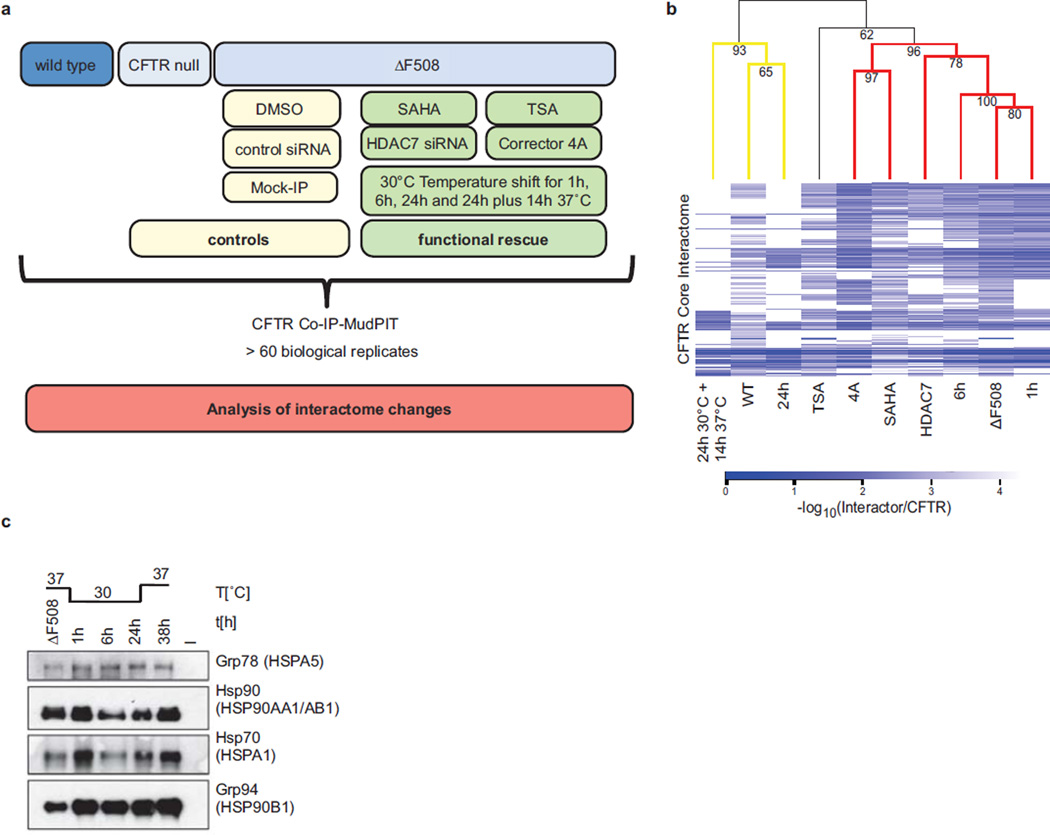
Extended Data Figure 3.
Overview of drug treatment, siRNA mediated knockdown and temperature shift experiments. a. Schematic showing the experimental outline. b. Expression of different heat shock proteins. The Western blot shows expression of HSP90 (encoded by HSP90AA1 and HSP90AB1), GRP78 (HSPA5), GRP94 (HSP90B1) and HSP70 (HSPA1) during temperature shift to 30°C. c. CID MS2 spectrum of the acetylated HSPA1 peptide LDK(42.01)AQIHDLVLVGGSTR acquired on an Orbitrap XL. Mass shift of 42.01 indicates acetylation of HSP70 at Lysine 328 in the ATPase domain. Depicted are b- and y-fragment ions with the corresponding m/z values. All data are from independent biological replicates, wt (n=7), ΔF508 (n=8), SAHA (n=4), TSA (n=4), HDAC7 (n=3), Cmpd 4a (n=3). d. Hierarchical clustering analysis of the CFTR core interactomes shows that the ΔF508 CFTR interaction profile clusters with high significance with those of ΔF508 CFTR at 1 h and 6 h temperature shifts to 30°C (mutant cluster), whereas temperature shift to 30°C for 24 h and temperature shift to 30°C for 24 h with reversal cause the respective ΔF508 CFTR interaction profiles to significantly cluster with that of wt CFTR. Bootstrap values (10,000 samplings) are given for each tree node. Significant (bootstrap value > 90, yellow) and highly significant clusters (bootstrap value > 95, red) are coloured on the dendrogram. The heatmap indicates the relative protein abundance values measured by mass spectrometry as negative log10 ratios of interactors relative to CFTR. White in the heatmap indicates that no interaction was observed.