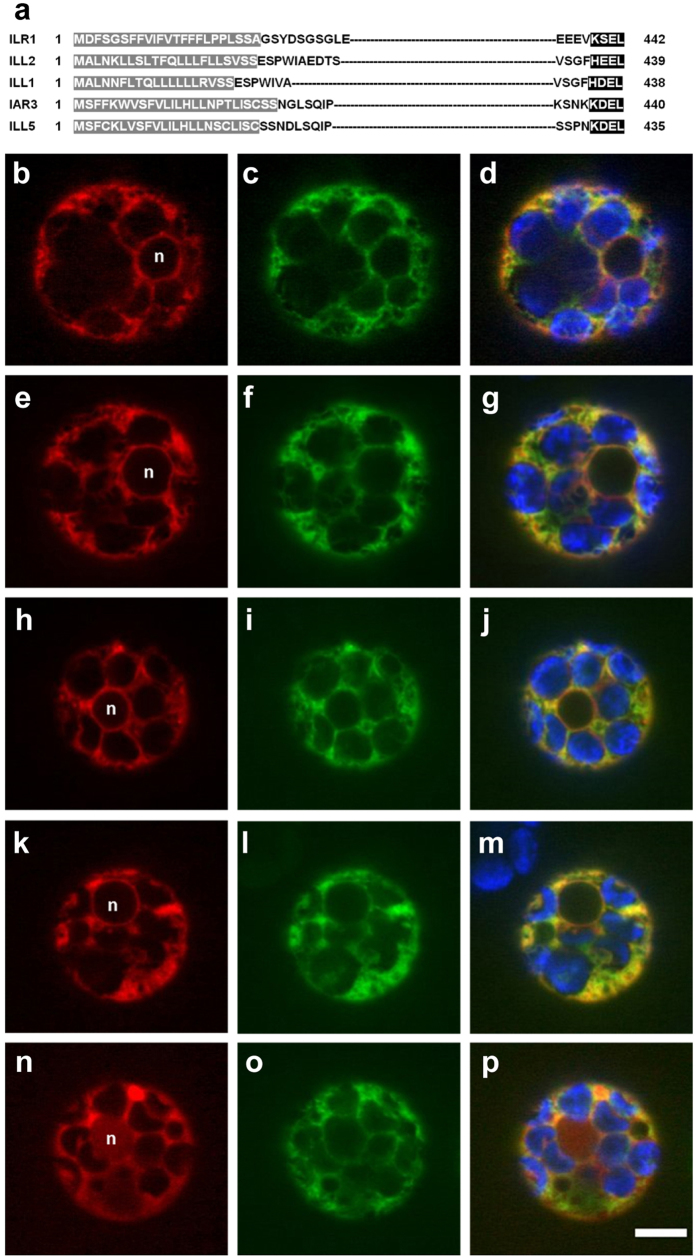
Figure 4. Subcellular localization of ILR1, ILL2 and IAR3.
(a) N- and C-terminal protein sequences of Arabidopsis ILR1, ILL2, IAR3, ILL1 and ILL5. The predicted signal peptides, according to TargetP40, are highlighted in grey; the potential retention signals are highlighted in black. (b–j) Subcellular localization of ILR1, ILL2 and IAR3. Confocal images of Arabidopsis leaf protoplasts co-expressing the mCherry-HDEL ER marker (b,e,h) and either ILR1 (c) ILL2 (f) or IAR3 (i) fused to eGFP. (k–p) Co-localization analysis of ILL2 depleted of the potential retention signal HEEL (l) with the ER mCherry marker (k) or ILR1 depleted of the potential retention signal KSEL (n) with ILR1 fused to eGFP (o). Merged signal of red and green channels (d,g,j,m,p) indicates co-localization. In the merged images the chloroplasts are visualized in blue and in the red channel images the nucleus position is indicated (n). The scale bar corresponds to 5 μM.