Fig. 1.
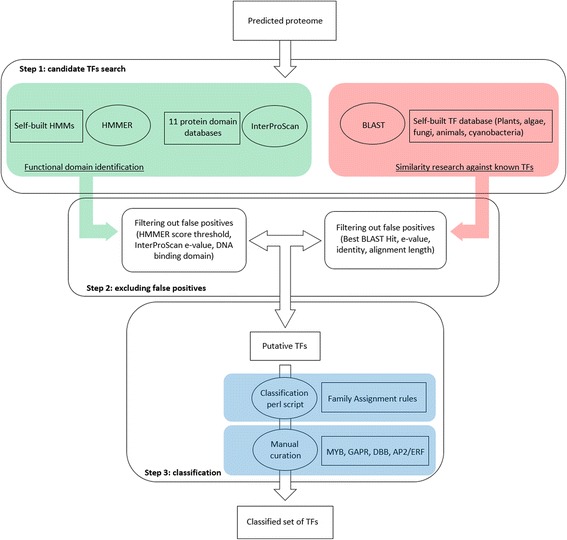
Identification pipeline. The pipeline is divided into three steps. Step One uses two strategies: i) a similarity search against an algae-based self-built database of known TFs with BLAST software; ii) functional domain annotation with InterProScan and HMMER software. The protein list obtained is the subject of the Step Two: the filtration of false positives according to specific parameters (see Methods). The last step consists in the classification of the putative TF list obtained in Step Two using a homemade perl script followed by manual curation for specific cases (see Methods)
