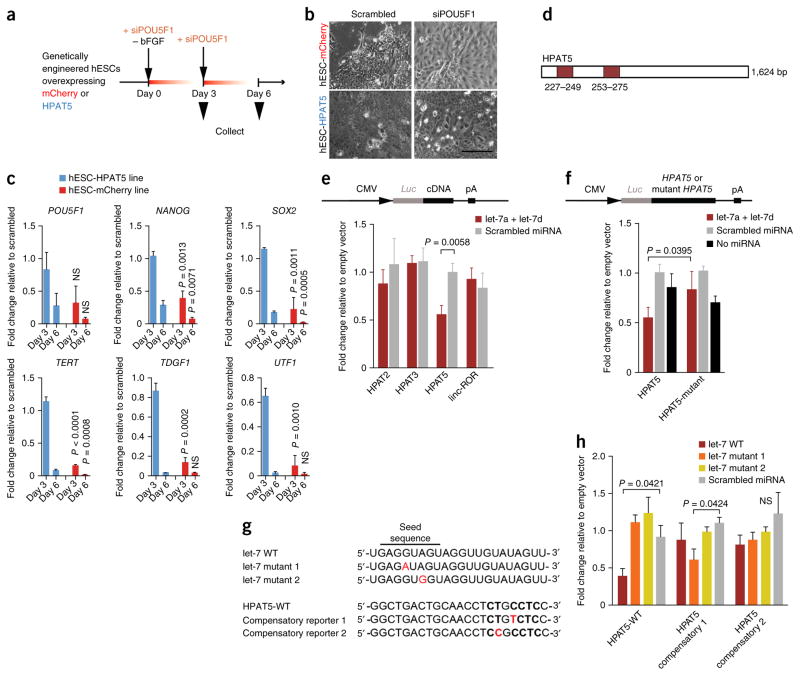
Figure 4.
HPAT5 binds directly to let-7. (a–c) HPAT5-OE hESCs suppress differentiation mediated by siRNA to POU5F1 and bFGF removal. (a) Outline of the protocol. (b,c) The expression of key pluripotency markers decreases with delay in comparison to the mCherry-OE control line. P values are calculated for the comparison of the mCherry-OE and HPAT5-OE lines on the same days (c), with this evidence supported by morphological changes (after day 3) (b) (n = 2 independent experiments). Scale bar, 50 μm. (d) Two predicted let-7 binding positions in HPAT5 identified by RegRNA 2.0 (miRanda). (e,f) Target validation using luciferase reporters in HEK293 cells. The relative luciferase activity (shown as fold change relative to empty vector) was assayed 48 h after cotransfection of cells with the indicated miRNAs or control (scrambled miRNA) together with wild-type reporter (e) or the let-7 miRNAs together with wild-type or mutant reporter (f). (g) The point mutations (in red) introduced into two mutant let-7 mimics and two mutant HPAT5 reporters for compensatory analysis. WT, wild type. (h) Analysis of the effects of the point mutations using luciferase reporters in HEK293 cells. Relative luciferase activity (shown as fold change relative to empty vector) of wild-type or mutant reporters was assayed 48 h after cotransfection of cells with the indicated miRNAs or scrambled miRNA. Representative results from n = 2 (c,e,f,h) independent experiments; n = 3 samples; data are shown with s.e.m.