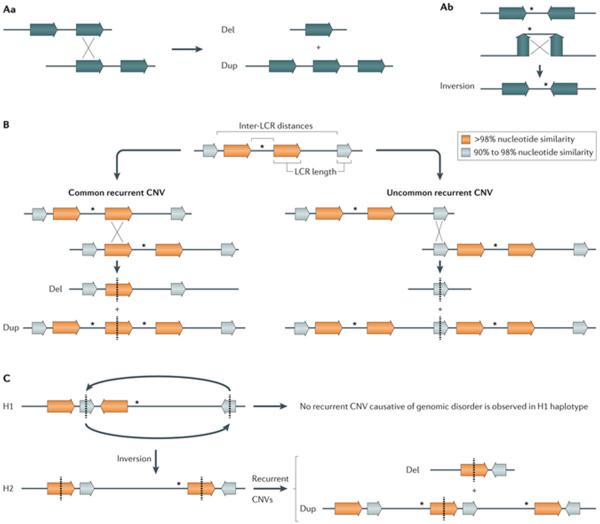
Figure 2. Nonallelic homologous recombination mechanism.
A | Nonallelic homologous recombination (NAHR) between directly oriented repeats generates deletions and duplications (part Aa), whereas NAHR between inverted repeats generates the inversion of the segment in between the repeats (part Ab). B | The rate of NAHR may vary within a particular locus owing to the presence of low-copy repeat (LCR) pairs with distinct susceptibility risk. For example, NAHR rate positively correlates with LCR length and inversely correlates with inter-LCR distance, which can lead to common recurrent and uncommon recurrent copy number variants (CNVs) at a given locus identified in different patients with a genomic disorder30 (for example, in common recurrent and uncommon recurrent deletions and reciprocal duplications in Smith–Magenis syndrome and Potocki–Lupski syndrome, respectively)46. C | NAHR rate may vary among individuals with distinct genomic architecture in a given locus or a structural variant haplotype that results in directly oriented LCR. This is exemplified by inverted structural haplotypes that present with distinct risks of undergoing NAHR owing to structural variability in LCR content and orientation. For instance, two divergent structural variant haplotypes, H1 and H2, are observed in 17q21.31 region but deletions that cause Koolen-De Vries syndrome (OMIM 610443)150 occur on chromosomes carrying the H2 haplotype. Dosage-sensitive genes flanked by LCRs are represented by asterisks. LCRs are represented by horizontal colour-matched arrows.