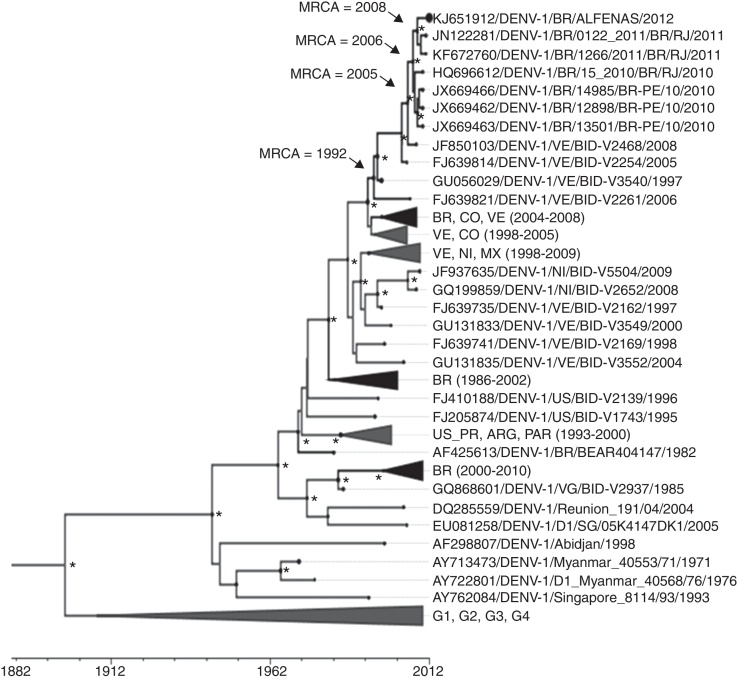
Fig. 2.
Phylogenetic analysis of DENV-1 strains. Bayesian coalescent analysis of DENV-1 strains was performed using a total of 119 envelope protein coding sequence (1485 bp). The phylogeny and time of the most recent common ancestor (MRCA) were estimated using the year of isolation of each DENV-1 strain as the calibration point, under the relaxed molecular clock, with the Tamura Nei Model, with discrete Gamma distribution. The maximum clade credibility tree is shown, summarizing data from three independent runs. Posterior probabilities with values ≥98 are represented by an asterisk (*) and black circles in each node or branch represent the nucleotide substitution rate. The years that the some most recent common ancestors (MRCA) were estimated to exist are shown for some nodes. Sequences are initially identified by their GenBank accession number. Brazilian strains or lineages containing Brazilian strains are shown in black. For clarity purposes some branches were collapsed as follows: [BR, CO, VE (2004–2008): FJ850093, FJ639820, FJ850100, FJ639818, GQ868562, FJ639823, FJ882579, FJ639813, FJ639808, FJ639797, GQ868570, GQ868568, GU131949, GQ868569]; [BR (1986–2002): JX669475, KF672764, FJ850073, KF672763, AY277665, JX669470, JX669471, JX669474, JX669473, JX669472, JX669468, JX669467, AF311958, AF311957, JX669469, AF311956, KF672761, KF672762, AF226685, JN122280, HQ026760, AF425614]; [BR (2000–2010): HM043709, HQ026762, HQ696614, HQ026761, JX669464, GU131863, KF672759, FJ850087, JX669461, JX669465, FJ850081, FJ850075, FJ850070, FJ850090, FJ850084, FJ850077, FJ850071]; [VE, CO (1998–2005): GU131834, GU131948, FJ639743, GQ868561, GU131837, FJ639740, GQ868560]; [VE, NI, MX (1998–2009): JQ287666, FJ024479, FJ182002, GQ868530, FJ024485, FJ547088, EU596501, FJ898437, FJ898433, GQ868499, FJ810419, GU131984, GQ868500, HQ166037, FJ024483, GU131957, GU056032]; [US_PR, ARG, PAR (1993–2000): AF514878, AY277666, AY277664, FJ390380, FJ410184, EU482567] and [G1, G2, G3, G4: FJ196845, FJ196842, DQ285561, JF960217, EU863650, DQ672563, EU848545, EU081262]. BR: Brazil, VE: Venezuela, CO: Colombia, NI: Nicaragua, MX: Mexico, US_PR: Porto Rico, ARG: Argentina, PAR: Paraguay, G1, G2, G3, G4: genotypes 1, 2, 3 and 4, respectively. Analysis was carried out using BEAST package v.1.8.1, BEAUTi v.1.8.1, Tracer v.1.6.0, TreeAnotator v.1.8.1 and FigTree v.1.4.2 and LogCombiner v1.8.1.