Abstract
BACKGROUND
Over the past 2 years, noninvasive prenatal testing (NIPT), which uses massively parallel sequencing to align and count DNA fragments floating in the plasma of pregnant women, has become integrated into prenatal care. Professional societies currently recommend offering NIPT as an advanced screen to pregnant women at high risk for fetal aneuploidy, reserving invasive diagnostic procedures for those at the very highest risk.
CONTENT
In this review, we summarize the available information on autosomal and sex chromosome aneuploidy detection. Clinical performance in CLIA-certified, College of American Pathology–accredited laboratories appears to be equivalent to prior clinical validation studies, with high sensitivities and specificities and very high negative predictive values. The main impact on clinical care has been a reduction in invasive procedures. Test accuracy is affected by the fetal fraction, the percentage of fetal DNA in the total amount of circulating cell-free DNA. Fetal fraction is in turn affected by maternal body mass index, gestational age, type of aneuploidy, singleton vs multiples, and mosaicism. Three studies comparing NIPT to serum or combined screening for autosomal aneuploidy all show that NIPT has significantly lower false-positive rates (approximately 0.1%), even in all-risk populations. A significant number of the discordant positive cases have underlying biological reasons, including confined placental mosaicism, maternal mosaicism, cotwin demise, or maternal malignancy.
SUMMARY
NIPT performs well as an advanced screen for whole chromosome aneuploidy. Economic considerations will likely dictate whether its use can be expanded to all risk populations and whether it can be applied routinely for the detection of subchromosome abnormalities.
Noninvasive prenatal testing (NIPT) refers to the sequence analysis of the cell-free DNA fragments that circulate within the blood of pregnant women. The term is generally used in the context of prenatal screening for autosomal aneuploidy. The focus of this review is on the lessons that have been learned since NIPT became clinically available in 2011, and specifically what they mean for the future widespread adoption of this technology. The automotive metaphor alluded to in the title of the review is especially appropriate, given that this field moves fast.
NIPT for Autosomal Aneuploidy
The seminal observation that cell-free fetal (cff) DNA fragments could be isolated and analyzed from the blood of pregnant women was made in 1997 (1). Although it is called fetal DNA, it derives from apoptotic cells in the placenta (2, 3). The DNA isolated from maternal blood is a mixture of fetal and maternal DNA in varying proportions that change as pregnancy progresses. A fundamental advance that occurred in 2007 was the concept that aneuploidy detection required a 2-step approach of shotgun sequencing of DNA followed by counting statistics (4, 5). Subsequent studies from the same groups demonstrated proof of principle in plasma samples from pregnant women carrying aneuploid fetuses (6, 7). Between 2010 and 2012, multiple clinical trials were performed that essentially showed similar performances in the sensitivity and specificity of detection of trisomies 21 and 18 (8–18). These trials, and their published results in peer-reviewed journals, led to the commercial release of NIPT for autosomal aneuploidy in 2011. More than 100 000 clinical tests have been performed to date in the US. The test is offered between 10 and 40 weeks of gestation.
TECHNICAL APPROACHES TO NIPT
Sequencing methods
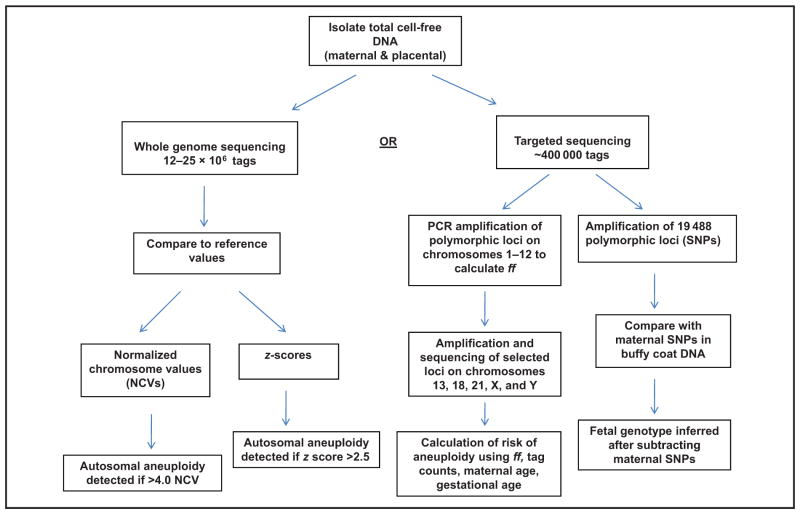
Three different approaches are currently used in clinical practice: massively parallel sequencing (MPS) of the whole genome, targeted sequencing, and single nucleotide polymorphisms (SNPs) (Fig. 1). All approaches sequence the total (maternal and fetal) cell-free DNA that is isolated from maternal plasma, which is fragmented. Fetal DNA fragments are predominantly 143 bp in length, and the maternal DNA fragments are generally 166 bp (19). With MPS, only the first 25 or 36 bp of each fragment is sequenced; this sequence is unique enough to allow alignment to a specific physical location within the human genome. A mapped sequence is known as a tag. The companies that perform whole-genome sequencing map between 12 and 25 million tags per sample. The number of tags at a specific chromosome is then counted and compared to reference values obtained from a normal human genome. An excess or deficiency in the number of counts, expressed as a z-score or normalized chromosome value (NCV), implies aneuploidy for that chromosome. It is important to emphasize that the chromosome counts derive from both maternal and fetal DNA fragments, and they are treated equally statistically. Any deviation from the reference values could be maternal or fetal in origin.
Fig. 1.
Major differences in the technical approaches to sequencing of circulating cell-free DNA in the plasma of pregnant women.
Targeted sequencing limits mapping of tags to the chromosomes of clinical interest (e.g., 13, 18, 21, X, and Y). This allows a significant reduction in the overall number of tags required, with a consequent reduction in cost. With targeted sequencing, an initial amplification of 192 loci on chromosomes 1 through 12 that contain SNPs is performed (16). At loci where the fetal alleles differ from the maternal alleles, a maximum likelihood estimate by use of the binomial distribution of markers is applied to calculate the fetal fraction, which is included in the assessment of whether aneuploidy is detected or not.
The SNP approach selectively amplifies and sequences 19 488 polymorphic loci on chromosomes 13, 18, 21, X, and Y (20, 21). The algorithm used in this approach incorporates maternal genotype information and recombination frequencies to construct billions of theoretical fetal genotypes. The software calculates a relative likelihood for each hypothetical genotype by comparing it with data obtained from the maternal buffy coat DNA sequence. The SNP approach does not require reference values. It also avoids some of the technical problems associated with guanine cytosine (GC) bias (discussed below) and can detect fetal triploidy. However, because of the need to compare sequences with the maternal genotype, it cannot be used in egg donor pregnancies.
Counting statistics
Fan and Quake (22) showed that sequencing results were biased in chromosomes with increased or decreased GC base content. They described a computational method to remove the GC bias. This eliminated the variance in the distribution of sequence tag counts and increased statistical confidence in the detection of aneuploidy. They also showed that with increased sequence tags there was increased precision of calling a sample aneuploid (22). After GC correction, MPS counting statistics follow a normal distribution. Each testing company has different proprietary algorithms to determine cutoffs between normal and abnormal. The classification cutoff will determine the expected false-positive rates.
BIOLOGICAL FACTORS THAT AFFECT ACCURACY OF NIPT
The accuracy of NIPT is affected by multiple technical and biological factors that are all integrated with and affected by each other. These include the number of sequencing tags, fetal fraction, GC base content, and CV.
Fetal fraction
The so-called fetal DNA is really from the placenta (23, 24). cff DNA is detectable even in the absence of an embryo (23). Faas et al. (25) described a case in which the plasma DNA and amniocentesis results were 45,X. Cytogenetic studies of the placenta showed that the mesenchymal core had a 46,XX karyo-type and the cytotrophoblast layer was 45,X. This study is often cited as the basis for the fact that cff DNA derives from cytotrophoblasts.
Fetal fraction (ff) is the amount of cff DNA divided by the total DNA, which comprises fetal plus maternal cell-free DNA. Circulating cell-free DNA in healthy adults derives mainly from hematopoietic cells undergoing apoptosis (26). In obese pregnant women, however, it partly derives from apoptosis and necrosis of adipose tissue and stromal vascular cells (27).
The ff can be computed on the basis of polymorphic sequence variation (17) or methylation differences (28) between maternal and fetal DNA. If the fetus is male or aneuploid, the ff can be calculated by use of sequence tag data from the X or aneuploid chromosomes (29). The advantage of the latter approach is that is does not require a separate processing step. The disadvantage is that it cannot be used for euploid female fetuses without deeper sequencing.
Most of the current serum markers used in Down syndrome screening follow a gaussian curve. In a reanalysis of the distribution of the percent of chromosome 21 markers in 212 fetuses affected with trisomy 21 (18), Canick et al. (30) found that the distribution was not gaussian; it was mainly influenced by ff. The higher the ff, the easier it is to call aneuploidy, because the z-scores are higher. Similarly, low ffs (<4% in some clinical laboratories) are related to test failures and false-negative test results. ff is affected by both maternal and fetal factors (Table 1).
Table 1.
Factors that affect fetal fraction of circulating DNA.
| Positive correlation | No correlation |
|---|---|
| Maternal weight (29–31, 33) | Maternal age (29, 32) |
| Gestational age (29, 33) | Nuchal translucency measurement (31, 32) |
| Maternal ethnicity (32) | Maternal ethnicity (29, 31) |
| Serum PaPP-A and free βhCG levels (31, 32)a | Plasma/serum storage time (31) |
| Fetal karyotype (↓ in trisomy 21, ↓ in trisomy 18) (29, 32) | Fetal sex (31, 32) |
| Maternal smoking (32) | Maternal smoking (31) |
PaPP-A, pregnancy-associated plasma protein A; βhCG, β human chorionic gonadotropin.
Maternal body mass index
Maternal body mass index (BMI) is the most significant demographic variable that affects ff. This has been consistently demonstrated in every study in which its effect has been analyzed (12, 30–33). In a study of pregnant women at 11–13 weeks of gestation, Ashoor et al. (32) showed that the median ff was 11.7% in women who weighed 60 kg; this decreased to 3.9% in women who weighed 160 kg.
Why do heavier pregnant women have lower ffs? Partially this is due to a dilution effect from a larger circulatory volume (30). In addition, remodeling of adipose tissue in obese pregnant women is associated with a 2-fold increase in GAPDH (a marker of total DNA) in serum (27). Total circulating cell-free DNA is proportionately increased in pregnant women as a function of BMI (34, 35). The fetal DNA is unaffected, but the increased maternal DNA results in an overall lower ff.
Gestational age
In a large study (n = 22 384), Wang et al. (33) demonstrated that ff increased by 0.1% per week between 10 and 21 weeks of gestation (P < 0.0001). After 21 weeks, ff increased by 1% per week (P < 0.0001). In that study, 1.9% of samples were redrawn because the initial ff was too low for accurate analysis. In 76 of 135 samples (56%) in which a repeat sample was obtained from the same individual, there was a >4% ff in the second sample. Together, maternal weight and fetal gestational age accounted for as much as 27% of the interindividual variation seen in ff (33).
Fetal aneuploidy
Using MPS tag counts, Rava et al. (29) compared ffs in fetuses with trisomies 21, 18, and 13 and monosomy X to euploid male fetuses. Relative to euploid males, fetuses with trisomy 21 have an increased ff, and fetuses with trisomy 18, 13, and monosomy X have a decreased ff. Similar results for trisomies 21 and 18 have been reported by different methods (31, 32). The higher ff in fetuses with trisomy 21 may be 1 reason that test performance for trisomy 21 is better than for trisomies 18 and 13.
Singleton vs multiple gestations. In an initial observation, Sehnert et al. (10) reported on the use of MPS in 5 twin pregnancies. All fetuses were correctly called. In a larger study of 25 twin pregnancies, total ff was significantly higher (mean 18.1%) compared with singletons (mean 13.4%). All aneuploid pregnancies were correctly classified, with a median chromosome 21 z-score of 12.3 (36).
If both twins have the same genotype, they can effectively be treated like a singleton gestation, and the z-scores for the aneuploid chromosome should have a linear relationship with ff. The difficulty arises when twins are discordant for aneuploidy, because ff is not doubled. Although the overall ff is increased, the ff per fetus is reduced compared with singletons. With sequence tag counting data from multiple gestations in which at least 1 fetus had a genotype that differed from the mother’s (e.g., male or aneuploid), Srinivasan et al. (37) showed that the ff per fetus was reduced. They calculated that in approximately 10%–15% of cases, the ff would be lower than 4%, and therefore there would be a risk of a false-negative result.
To quickly determine if a twin gestation was dizygotic, the group at the Chinese University of Hong Kong used a proprietary algorithm (38) to identify regions of the genome in which the maternal and fetal sequences differed. They then calculated the average apparent ff for every 1000 informative SNPs on contiguous blocks of DNA within the targeted areas of the genome (39, 40). Results were confirmed by use of chorionic villi or umbilical cord blood. In 2 dizygotic twin cases (each discordant for aneuploidy), large regional fluctuations in ff were observed, consistent with dizygosity. This method allows twin zygosity to be successfully verified and facilitates an estimate of whether there is adequate fetal DNA present per fetus.
Mosaicism
If there is placental or fetal mosaicism with a cell line that has autosomal aneuploidy, a higher overall ff will increase the chance that the partial DNA contribution from a mosaic abnormal line will be found (30). Similarly, a high percentage of abnormal cells will increase the chance that sequences from the aneuploid chromosome will be detected. In the results from the MELISSA trial, all mosaic cases with at least 29% of cells positive for trisomies 18 and 21 were detected as aneuploid (14). In contrast, low percentages of mosaicism (10%) or low effective ffs have led to false-negative results (30).
Chromosome biology
Chromosomes that have GC content in the midrange (such as 21, 18, and X) perform best with MPS (7). Chromosome 13 has one of the lowest GC contents. For this reason, specific approaches have been used to normalize the GC ratios for chromosome 13, which have resulted in increased sensitivity of detection of trisomy 13 (22, 41). An additional factor that affects sensitivity is the CV in the sequencing results for a specific chromosome. A smaller CV results in larger z-scores, making it easier to separate aneuploid from euploid samples. Chromosome 18 has approximately half of the CV observed for chromosomes 13, 21, and X (29). The clinical implication is that trisomy 18 can be detected at lower ffs. It is therefore easier to detect low-grade mosaicism, which may be 1 reason why there are more discordant results between NIPT and karyotype with trisomy 18 (42).
Clinical and Laboratory Experience in the “Real World”
As NIPT has transitioned from clinical trials to clinical care, reports are beginning to be published on screening laboratory performance in the clinical setting. For example, Futch et al. (43) described an initial 9-month experience in their CLIA-licensed, College of American Pathology–accredited laboratory. Maternal plasma samples were sequenced if they were drawn into Streck tubes (44), accompanied by a test requisition form and signed patient informed consent, received within 5 days of blood draw, and from singleton pregnancies. Of the 5974 tests ordered, 43 (0.7%) were cancelled for technical reasons. Reports were issued for each individual chromosome tested (e.g., 13, 18, 21, X). In 2.8% of samples, at least 1 of the chromosomes had an unclassifiable result. The mean turnaround time was 5.1 business days. Aneuploidy was detected in 4.8% of samples, which suggested that testing was being performed on a high-risk population as is currently recommended by most professional societies (45–47). Of the aneuploid cases, one third were confirmed by karyotype, an additional half were consistent with other clinical information, and the remainder were in ongoing pregnancies. In the cases for which there was clinical outcome information, there were 5 false-negative cases (2 trisomy 21, 2 trisomy 18, 1 monosomy X), for a rate of 0.08%. There were 14 (0.2%) false-positive cases that were discordant with the fetal karyotype. Importantly, many of these cases had underlying biological reasons for the excess sequences detected, such as confined placental mosaicism (CPM), maternal sex chromosome aneuploidy, maternal malignancy, low-level fetal mosaicism, and cotwin demise (Table 2). This report demonstrated that in a clinical setting, NIPT performance met or exceeded characteristics established in the prior clinical validation studies (43). In the vast majority of samples, there was no aneuploidy detected. The negative predictive value was 99.6% for trisomy 21.
Table 2.
Potential explanations for NIPT fetal karyotype discordance.
| NIDT result | Fetal karyotype | Explanation | Reference |
|---|---|---|---|
| Autosomal aneuploidy | |||
| Trisomy 13 | 46,XX/46,XX, +13, der (13;13) (q10;q10) | Confined placental mosaicism | Mennuti et al. (42) |
| Trisomy 13 | 46,XY | Confined placental mosaicism | Mennuti et al. (42) |
| Trisomy 13 | 46,XY [12]/47,XY,+13 [10] | Confined placental mosaicism | Hall et al. (53) |
| Trisomy 21 | 46,XX, upd (21) mat | Confined placental mosaicism/Rescue disomy | Pan et al. (54) |
| Trisomy 22 | Normal by sequencing | Confined placental mosaicism | Choi et al. (56) |
| Trisomy 18 | 46,XX | Maternal mosaicism 46, XX[47]/47, XX +18[13] | Song et al. (49) |
| Trisomy 13 (2 cases) | Unknown | Cotwin demise | |
| Trisomy 21 and monosomy X | Unknown | Cotwin demise | Futch et al. (43) |
| Trisomy 21 | Euploid | Laboratory error | Futch et al. (43) |
| Trisomy 13, monosomy 18 | 46,XY | Maternal tumor | Osborne et al. (58) |
| SCA | |||
| 47,XXY | 49,XXX, +7, +21 | Confined placental mosaicism | Lau et al. (55) |
| 47,XXX | 46,XX | Maternal nonmosaic SCA 47,XXX | Yao et al. (57) |
| 45,X | 46,XX | Maternal SCA mosaicism; 45,X[3]/46,XX[27] | Lau et al. (55) |
Although in the US and Canada there are clear professional recommendations to limit NIPT to pregnancies at high risk for aneuploidy (45–47), studies are starting to emerge on test performance in the so-called all-risk pregnancies. In 1 report from a private obstetric practice in Atlanta, Fairbrother et al. (48) offered NIPT and first-trimester combined screening to all pregnant women carrying singleton gestations (n = 289). NIPT results were obtained in 98.6% of samples, and the mean time from blood draw to test result was 9.3 calendar days. At a cutoff value of 1:311, 12 of 284 first-trimester screens were called positive (4.5%). With NIPT, all 284 samples were considered to be low risk (<1:10 000) for trisomies 13, 18, and 21. One woman requested a definitive fetal karyotype; the fetus was euploid. On the basis of the significantly decreased false-positive rate with NIPT, this practice now offers NIPT as their primary aneuploidy screen to all women with singleton pregnancies after 10 weeks.
In China, investigators compared NIPT to second-trimester serum triple screening in 1916 low-risk pregnant women ages 20–34 years (49). There was a 3.8% test failure rate. At a cutoff risk of 1 in 270, 249 of 1741 (14.3%) women were triple-screen positive. Twelve of 1741 (0.68%) women had a positive NIPT result (11 cases of fetal aneuploidy and 1 discordant result for trisomy 18 due to maternal mosaicism). The sensitivity of detection of autosomal aneuploidy by NIPT was 100% (11 of 11), the specificity was 99.4% (1729 of 1730), and the positive predictive value (PPV) was 91.67% (11 of 12). In contrast, serum triple screening had a sensitivity of 54.5% (6 of 11), specificity of 85.9% (1487 of 1730), and PPV of 2.4% (6 of 249). These authors concluded that NIPT performance was effective in a low-risk population and outperformed second-trimester serum screening.
Another recent implementation study performed at a center in the UK compared NIPT at 10 weeks with the combined test at 12 weeks in 1005 singleton pregnancies (50). Although this was described as an all-risk study, the mean maternal age was 36.7, and only 85.7% of women conceived spontaneously. Test results were available in 95.2% of cases. The 4.8% (48 of 1005) of test failures were due to problems with delivery to the laboratory, a low ff, or test assay failure. In 40 of these 48 cases, a repeat blood sample provided results approximately two thirds of the time. There were 11 cases of trisomy 21, 5 cases of trisomy 18, and 1 case of trisomy 13. Fifteen of 16 were confirmed by CVS. There was 1 false-positive case of trisomy 18. The false-positive rates were 0.1% (1 of 968) for NIPT and 3.4% (33 of 968) for the combined test. These authors also concluded that the main advantage of DNA testing is the substantial reduction in false positives and a clear separation of cases at high and low risk for aneuploidy. The disadvantages of NIPT were the significant percentage of test failures and the need to follow up on abnormal results by invasive testing.
These 3 studies show that NIPT performs better than serum screening or the combined test, even in all-risk populations. The current cost of NIPT, however, is too high to offer to everyone. Investigators are beginning to explore a contingent approach, in which the results of the combined test are used to identify a population of pregnant women who could benefit from NIPT as a secondary screen (51). This approach would reduce costs by decreasing invasive procedures and eliminating some of the sequencing expenses. The disadvantage of applying NIPT to only the screen-positive cases would leave the original screen-negative cases untested, which could miss approximately 10%–15% of the trisomy 21 fetuses.
FALSE POSITIVES AND NEGATIVES
The false-positive rates for NIPT are on the order of 0.1%–0.2%. What remains unknown, however, is what portion of the discordancy between NIPT and fetal results is attributable to the demarcation between euploidy and aneuploidy established by the statistical algorithms, and how much is due to biologic explanations uncovered by the greater sensitivity of MPS.
CPM is present in 1%–2% of first-trimester placentas (52). Several instances of CPM detected at CVS have been reported in association with discordant positive NIPT results (Table 2). Mennuti et al. (42) noted 2 cases of CPM in association with noninvasive detection of trisomy 13. In the first case, trisomy 13 was detected only in the CVS direct sample. In the second case, the CVS direct preparation revealed 46,XX, +13, der (13, 13)(q10;q10), consistent with the NIPT results. Hall et al. (53) described NIPT results with an increased risk for trisomy 13. CVS demonstrated trisomy 13 mosaicism, by both FISH and long-term culture (47,XY, +13[10]/46,XY) (12). At delivery, the newborn was growth restricted, but had no features of trisomy 13. His blood karyotype was 46,XY. Additionally, the placenta had evidence of trisomy 13 mosaicism in 2 of 4 sites sampled.
Of further interest is the report describing uniparental disomy (UPD), the presence of both chromosomes inherited from 1 parent as the result of disomic rescue (54). In a case of NIPT positive for trisomy 21, quantitative fluorescent (QF)-PCR of a CVS sample at 14 weeks’ gestation suggested isodisomy type iUPD21 on the basis of 7 short tandem repeat (STR) markers all inherited from the mother. After pregnancy termination, QF-PCR of 4 placental biopsies revealed 1 with disomy (the original iUPD21) and trisomy 21 in the remaining 3 sites.
CPM has also been reported for other chromosomes. In a large Chinese population study, a single NIPT case result suggested mosaicism for multiple aneuploidies: 47,XXY, trisomy 21, and trisomy 7 (55). The CVS long-term culture karyotype was 49,XXX, +7, +21[24]/46,XY (6). In another case, sequencing results suggested trisomy 22; this led to the identification of trisomy 22 in 3 placental biopsy sites with disomy 22 in the cord blood. At delivery, the infant was growth restricted (3%) but had no dysmorphic features (56).
Maternal conditions should also be considered as possible biologic explanations of NIPT-fetal discordant results. These include constitutional maternal aneuploidies, most likely involving sex chromosomes, and conditions with intrinsic genomic alterations, such as solid tumors (Table 2).
The initial report of a maternal sex chromosome aneuploidy (SCA) occurred in a completely normal 25-year-old pregnant woman whose NIPT results were consistent with triple X (57). Amniocentesis revealed a fetal 46,XX karyotype; her infant was normal. Maternal blood karyotype showed full 47,XXX. Her presentation emphasizes the wide range of phenotypic variability with the SCAs.
Maternal mosaicism for a SCA was also the underlying explanation in a 44-year-old woman whose NIPT results suggested abnormal X chromosome ratios that were inconsistent with the laboratory’s prior cases of fetal 45,X[55]. The maternal karyotype was 45,X[3]/46,XX[27]. The newborn’s karyotype was normal. SCAs, especially mosaic cases, are likely to be under-appreciated clinically. The possibility of their identification through NIPT requires discussion during pre-and posttest counseling.
Another source of maternal DNA that could result in discordant results is a solid tumor (58). In a pregnant woman, NIPT results at 13 and 17 weeks showed a double aneuploidy, trisomy 13, and monosomy 18. The fetal karyotype was 46,XY and a healthy male was born at term. Placental biopsies showed 46,XY at 6 sites, ruling out CPM. During a postpartum evaluation for pelvic pain, radiographs were notable for pathologic fractures involving the right superior and inferior pubic rami, as well as other changes suggestive of malignancy. Bone biopsy confirmed metastatic neuroendocrine carcinoma. Postpartum, the primary source was identified as a small cell carcinoma of vaginal origin. The majority of cells (80 of 100) from the tumor biopsy demonstrated an increased number of chromosome 13 signals by FISH relative to chromosome 18 signals, consistent with the NIPT results. The underlying rate of maternal cancers that present with discordant NIPT-fetal karyotype results is unknown, but warrants consideration when multiple aneuploidies are called.
With regard to false-negative cases, little information is available. The few reported cases suggest that the underlying explanation is either a sample mix-up (43), low ff (30, 43), or mosaicism (30).
NIPT FOR SCA
SCAs are present in 1 of 400 live births, a rate that is higher than the most common autosomal aneuploidies combined (59). NIPT for SCAs has been clinically available since 2012. SCAs were detected at lower sensitivities in the initial studies of high-risk women undergoing MPS for autosomal aneuploidy (14, 60). In a low-risk population of 1916 Chinese women, 2 of 4 SCAs were detected [50% sensitivity (95% CI 9.1%–90.1%), 100% specificity (95% CI 99.72%–100%)] (49).
Several challenges exist for the accurate quantification of the number of X chromosomes. These include the inherent sequencing bias associated with GC content of the X, the marked sequence similarity between the X and Y chromosomes, the small size of the Y chromosome that can lead to large variations in sequence assignments, and the presence of maternal or fetal mosaicism that can alter interpretation (61). A unique aspect of the X chromosome is inactivation, which is driven by specifically recognized histone variants, which may lead to variability in the X chromosome content of the cell-free DNA (62, 63).
In a training panel of 1546 cases, dual classification of the fetal sample by sex, followed by assessment of sex chromosome complement, provided an overall sensitivity of 100% (95% CI 82.3%–100%) and a false-positive rate of 0.1% (95% CI 0%–0.3%) (61). The test failure rate was 6% (95% CI 4.9%–7.4%). Application of this approach in a blinded study yielded a sensitivity of 96.2% (95% CI 78.4%–99.8%), false-positive rate of 0.3% (95% CI 0%–1.8%), and test failure rate of 5% (95% CI 3.2%–7.7%). Of the 185 female samples, 4 were interpreted as male. For 199 male samples, 1 was called as female and the remainder were correctly classified (191 XY, 5 XXY, 2 XYY).
Improved, second-generation sequencing has also been applied to the initial issues of lower sensitivity of SCA detection. Guex et al. (64) initially used a training panel and then a retrospective blinded study to develop a more refined approach that addressed GC sequencing bias in the sex chromosomes. They were able to correctly identify all cases of Turner syndrome (15 of 15) and 47,XXX (5 of 5), for a sensitivity and specificity of 100% (sensitivity 95% CI 79.9%–100%, specificity 95% CI 98.1%–100%). No test failures were reported.
In another study, SNP analysis was applied to 201 pregnancies (13 45,X; 2 47,XXY; 1 47,XYY; and 185 euploid samples). The sensitivity for monosomy X detection was 91.7% (95% CI 61.5%–99.8%), all 3 sex chromosome aneuploidies were called correctly, and there was 1 false-negative sex chromosome result called (63). There were no false-positive sex chromosome calls. However, of the 201 analyzed samples, 14 (7.0%) failed quality control. Despite these limitations, a SNP-based approach has the potential to address SCA detection in a way that circumvents the unique biology of the X chromosome.
Future Applications of Noninvasive DNA Testing
The most likely future clinical advance will be to extend from whole-chromosome to subchromosome abnormalities (65). Proof of principle that fetal subchromosomal abnormalities could be detected in maternal plasma DNA was established in 2 studies in which the fetal karyotypes were known a priori from invasive procedures. In the first, a paternally inherited 4.2-Mb deletion between 12p11.22 and 12p12.1 was demonstrated in maternal blood at 35 weeks of gestation (66). In the second, plasma DNA was sequenced from 2 women carrying fetuses with known 22q11.2 deletions (Di George syndrome) (67). In prospective blinded studies that were focused on detection of whole-chromosome aneuploidies, incidental findings of a microdeletion of 11q21–23 (10) and a duplication of 6q (14) were found.
More recently, research efforts have concentrated on approaches that would make the detection of subchromosome abnormalities feasible in a routine clinical setting. Chen et al. (68) described a bioinformatics algorithm that allows detection of large (approximately 10-Mb) deviations from the reference genome. From a set of 1311 samples with known fetal karyotype, they detected 4 abnormal cases: a 19-Mb deletion in 4p16–4p15.3, consistent with fetal Wolf–Hirschhorn syndrome; a double abnormality consisting of a 10-Mb deletion of 4q34.3–4q35.2 and a 17-Mb duplication of 7p22.3–7p21.1; and another double abnormality consisting of a 28-Mb duplication of 4q24.3 to qter and a 9-Mb deletion of 18p11.22 to pter. All of these cases were confirmed by karyotype. The fourth case, a duplication of 4q32.1 to 4q32.3, was not confirmed (a false positive). Applying this same algorithm, Lau et al. (55) demonstrated large deletions in 3 different fetuses. The advantage of this technique is that is does not require deeper sequencing.
Using computer simulations, Yu et al. (69) calculated the diagnostic sensitivity for detection of 3-Mb microduplications and deletions on a whole-genome survey. At a ff of 6%, there was 99% sensitivity with deeper sequencing. With 1.5 × 108 tags, they showed that they could detect 3 de novo microdeletions of 22q11.2, two 2.4-Mb microduplications of 22q11.2, and 1 double abnormality consisting of a 5.1-Mb microduplication of 3q29 and a 32.9-Mb microdeletion of 4q32.1–4q35.2. By partitioning the genome into bins of 1 Mb and 100 kb, Srinivasan et al. (70) were able to blindly detect 7 cases of fetal microdeletions, duplications, translocations, and a case of trisomy 20. One of the microdeletions detected was as small as 300 kb. In 4 cases with mosaic abnormalities, MPS did not detect the abnormal cell line, most likely because of the low ff. In 2 other cases in which the metaphase karyotype failed to identify the additional material, MPS identified both the translocation breakpoint and its chromosomal origin.
All of the 8 studies cited here suggest that it is possible to detect subchromosome abnormalities. This is beginning to be translated to clinical care, but widespread application will depend on cost of the assay and the sequencing depth.
Last, the “ultimate” prenatal test will be the noninvasive sequencing of the entire fetal genome (as reviewed in (71)). Although there have been 3 published studies to date that suggest that this is technically feasible (19, 72, 73), the clinical utility of such a test is unknown at present.
Summary
In this review, we have summarized the clinical experience over the past 2 years regarding the performance of NIPT for fetal aneuploidy. The data suggest that test performance is equivalent to or exceeds performance during the clinical validation studies performed worldwide. The high negative predictive values (>99%) associated with test performance have already translated to a significant reduction in invasive procedures (74). Despite initial concerns that the test might perform differently in all-risk populations of pregnant women (75), the evidence suggests otherwise (48–50). There are, however, still concerns that there are relatively high percentages of test failures. About a half to two thirds of test failures can be successfully resolved by drawing a second sample at a later time of gestation (33, 50). The ability to provide information on SCAs, later reflexing to fetal sex (without a medical indication), is a new frontier in prenatal screening that will require further ethical assessment.
As a final note, the rubber has hit the road in the high-performance race car we call NIPT. The pace of this field is unprecedented in clinical laboratory medicine. Of the references listed here, 50 of 75 (67%) have been published in the last 18 months. Although this makes it challenging to keep apprised of advances in the field, the generation of new knowledge will hopefully translate to advances in patient care equally rapidly.
Nonstandard abbreviations
- NIPT
noninvasive prenatal testing
- cff
cell-free fetal
- MPS
massively parallel sequencing
- SNP
single nucleotide polymorphism
- NCV
normalized chromosome value
- GC
guanine cytosine
- ff
fetal fraction
- BMI
body mass index
- CPM
confined placental mosaicism
- PPV
positive predictive value
- UPD
uniparental disomy
- QF
quantitative fluorescent
- STR
short tandem repeat
Footnotes
Author Contributions: All authors confirmed they have contributed to the intellectual content of this paper and have met the following 3 requirements: (a) significant contributions to the conception and design, acquisition of data, or analysis and interpretation of data; (b) drafting or revising the article for intellectual content; and (c) final approval of the published article.
Other Remuneration: D.W. Bianchi, Wiley-Blackwell Publishers.
Authors’ Disclosures or Potential Conflicts of Interest: Upon manuscript submission, all authors completed the author disclosure form. Disclosures and/or potential conflicts of interest:
Employment or Leadership: D.W. Bianchi, Verinata Health, an Illumina company.
Consultant or Advisory Role: D.W. Bianchi, Verinata Health.
Stock Ownership: None declared.
Honoraria: D.W. Bianchi, Verinata Health.
Research Funding: L.E. Wilkins-Haug, Arisoa, Sequenom–Women and Infants Hospital.
Expert Testimony: None declared.
Patents: None declared.
Role of Sponsor: The funding organizations played no role in the design of study, choice of enrolled patients, review and interpretation of data, or preparation or approval of manuscript.
References
- 1.Lo YM, Corbetta N, Chamberlain PF, Rai V, Sargent IL, Redman CW, et al. Presence of fetal DNA in maternal plasma and serum. Lancet. 1997;350:485–7. doi: 10.1016/S0140-6736(97)02174-0. [DOI] [PubMed] [Google Scholar]
- 2.Huppertz B, Kingdom JC. Apoptosis in the trophoblast-role of apoptosis in placental morphogenesis. J Soc Gynecol Invest. 2004;11:353–62. doi: 10.1016/j.jsgi.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 3.Hahn S, Huppertz B, Holzgreve W. Fetal cells and cell free fetal nucleic acids in maternal blood: new tools to study abnormal placentation? Placenta. 2005;26:515–26. doi: 10.1016/j.placenta.2004.10.017. [DOI] [PubMed] [Google Scholar]
- 4.Fan HC, Quake SR. Detection of aneuploidy with digital polymerase chain reaction. Anal Chem. 2007;79:7576–9. doi: 10.1021/ac0709394. [DOI] [PubMed] [Google Scholar]
- 5.Lo YM, Fun FM, Chan KC, Tsui NB, Chong KC, Lau TK, et al. Digital PCR for the molecular detection of fetal chromosomal aneuploidy. Proc Natl Acad Sci U S A. 2007;104:13116–21. doi: 10.1073/pnas.0705765104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fan HC, Blumenfeld YJ, Chitkara U, Hudgins L, Quake SR. Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proc Natl Acad Sci U S A. 2008;105:16266–71. doi: 10.1073/pnas.0808319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiu RW, Chan KC, Gao Y, Lau VY, Zheng W, et al. Noninvasive prenatal diagnosis of fetal chromosomal aneuploidy by massively parallel genomic sequencing of DNA in maternal plasma. Proc Natl Acad Sci U S A. 2008;105:20458–63. doi: 10.1073/pnas.0810641105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiu RW, Akolekar R, Zheng YW, Leung TY, Sun H, Chan KC, et al. Non-invasive prenatal assessment of trisomy 21 by multiplexed maternal plasma DNA sequencing: large scale validity study. BMJ. 2011;342:c7401. doi: 10.1136/bmj.c7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ehrich M, Deciu C, Zwiefelhofer T, Tynan JA, Cagasan L, Tim R, et al. Noninvasive detection of fetal trisomy 21 by sequencing of DNA in maternal blood: a study in a clinical setting. Am J Obstet Gynecol. 2011;204:205, e1–11. doi: 10.1016/j.ajog.2010.12.060. [DOI] [PubMed] [Google Scholar]
- 10.Sehnert AJ, Rhees B, Comstock D, de Feo E, Heilek G, Burke J, et al. Optimal detection of fetal chromosomal abnormalities by massively parallel DNA sequencing of cell-free fetal DNA from maternal blood. Clin Chem. 2011 doi: 10.1373/clinchem.2011.165910. [DOI] [PubMed] [Google Scholar]
- 11.Palomaki GE, Kloza EM, Lambert-Messerlian GM, Haddow JE, Neveux LM, Ehrich M, et al. DNA sequencing of maternal plasma to detect Down syndrome: an international clinical validation study. Genet Med. 2011;13:913–20. doi: 10.1097/GIM.0b013e3182368a0e. [DOI] [PubMed] [Google Scholar]
- 12.Sparks AB, Wang ET, Struble CA, Barrett W, Stokowski R, McBride C, et al. Selective analysis of cell-free DNA in maternal blood for evaluation of fetal trisomy. Prenat Diagn. 2012;32:3–9. doi: 10.1002/pd.2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palomaki GE, Deciu C, Kloza EM, Lambert-Messerlian GM, Haddow JE, Neveux LM, et al. DNA sequencing of maternal plasma reliably identifies trisomy 18 and trisomy 13 as well as Down syndrome: an international collaborative study. Genet Med. 2012;14:296–305. doi: 10.1038/gim.2011.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bianchi DW, Platt LD, Goldberg JD, Abuhamad AZ, Sehnert AJ, Rava RP, et al. Genome-wide fetal aneuploidy detection by maternal plasma DNA sequencing. Obstet Gynecol. 2012;119:890–901. doi: 10.1097/AOG.0b013e31824fb482. [DOI] [PubMed] [Google Scholar]
- 15.Ashoor G, Syngelaki A, Wagner M, Birdir C, Nicolaides KH. Chromosome-selective sequencing of maternal plasma cell-free DNA for firsttrimester detection of trisomy 21 and trisomy 18. Am J Obstet Gynecol. 2012;206:322, e1–5. doi: 10.1016/j.ajog.2012.01.029. [DOI] [PubMed] [Google Scholar]
- 16.Sparks AB, Struble CA, Wang ET, Song K, Oliphant A. Noninvasive prenatal detection and selective analysis of cell-free DNA obtained from maternal blood: evaluation for trisomy 21 and trisomy 18. Am J Obstet Gynecol. 2012;206:319, e1–9. doi: 10.1016/j.ajog.2012.01.030. [DOI] [PubMed] [Google Scholar]
- 17.Norton ME, Brar H, Weiss J, Karimi A, Laurent LC, Caughey AB, et al. Non-invasive chromosomal evaluation (NICE) study: results of a multicenter prospective cohort study for detection of fetal trisomy 21 and trisomy 18. Am J Obstet Gynecol. 2012;207:137, e1–8. doi: 10.1016/j.ajog.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 18.Dan S, Wang W, Ren J, Li Y, Hu H, Xu Z, et al. Clinical application of massively parallel sequencing-based prenatal noninvasive fetal trisomy test for trisomies 21 and 18 in 11 105 pregnancies with mixed risk factors. Prenat Diagn. 2012;32:1225–32. doi: 10.1002/pd.4002. [DOI] [PubMed] [Google Scholar]
- 19.Lo YM, Chan KC, Sun H, Chen EZ, Jiang P, Lun FM, et al. Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Sci Transl Med. 2010;2:61ra91. doi: 10.1126/scitranslmed.3001720. [DOI] [PubMed] [Google Scholar]
- 20.Nicolaides KH, Syngelaki A, Gil M, Atanasova V, Markova D. Validation of targeted sequencing of single-nucleotide polymorphisms for non-invasive prenatal detection of aneuploidy of chromosomes 13, 18, 21, X and Y. Prenat Diagn. 2013;33:575–9. doi: 10.1002/pd.4103. [DOI] [PubMed] [Google Scholar]
- 21.Zimmermann B, Hill M, Gemelos G, Demko Z, Banjevic M, Baner J, et al. Noninvasive prenatal aneuploidy testing of chromosomes 13, 18, 21, X and Y using targeted sequencing of polymorphic loci. Prenat Diagn. 2012;32:1233–41. doi: 10.1002/pd.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fan HC, Quake SR. Sensitivity of noninvasive prenatal detection of fetal aneuploidy from maternal plasma using shotgun sequencing is limited only by counting statistics. PLoS One. 2010;5:e10439. doi: 10.1371/journal.pone.0010439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alberry M, Maddocks D, Jones M, Abdel Hadi M, Abdel-Fattah S, Avent N, et al. Free fetal DNA in maternal plasma in anembryonic pregnancies: confirmation that the origin is the trophoblast. Prenat Diagn. 2007;27:415–8. doi: 10.1002/pd.1700. [DOI] [PubMed] [Google Scholar]
- 24.Masuzaki H, Miura K, Yoshiura K-I, Yoshimura S, Niikawa N, Ishimaru T. Detection of cell free placental DNA in maternal plasma: direct evidence from three cases of confined placental mosaicism. J Med Genet. 2004;41:289–92. doi: 10.1136/jmg.2003.015784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Faas BH, de Ligt J, Janssen I, Eggink AJ, de Wijnberger LD, et al. Non-invasive prenatal diagnosis of fetal aneuploidies using massively parallel sequencing-by-ligation and evidence that cell-free fetal DNA in the maternal plasma originates from cytotrophoblastic cells. Expert Opin Biol Ther. 2012;12(Suppl 1):S19–26. doi: 10.1517/14712598.2012.670632. [DOI] [PubMed] [Google Scholar]
- 26.Lui YY, Chik KW, Chiu RW, Ho CY, Lam CW, Lo YM. Predominant hematopoietic origin of cell-free DNA in plasma and serum after sex-mismatched bone marrow transplantation. Clin Chem. 2002;48:421–7. [PubMed] [Google Scholar]
- 27.Haghiac M, Vora NL, Basu S, Johnson KL, Presey L, Bianchi DW, et al. Increased death of adipose cells, a path to release cell free DNA into systemic circulation of obese women. Obesity. 2012;20:2213–9. doi: 10.1038/oby.2012.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nygren AO, Dean J, Jensen TJ, Kruse S, Kwong W, van den Boom D, et al. Quantification of fetal DNA by use of methylation-based DNA discrimination. Clin Chem. 2010;56:1627–35. doi: 10.1373/clinchem.2010.146290. [DOI] [PubMed] [Google Scholar]
- 29.Rava RP, Srinivasan A, Sehnert AJ, Bianchi DW. Circulating fetal cell-free DNA fractions differ in autosomal aneuploidies and monosomy X. [Epub ahead of print] Clin Chem. 2013 Oct 7; doi: 10.1373/clinchem.2013.207951. [DOI] [PubMed] [Google Scholar]
- 30.Canick JA, Palomaki GE, Kloza EM, Lambert-Messerlian GM, Haddow JE. The impact of maternal plasma DNA fetal fraction on next generation sequencing tests for common fetal aneuploidies. Prenat Diagn. 2013;33:667–74. doi: 10.1002/pd.4126. [DOI] [PubMed] [Google Scholar]
- 31.Ashoor G, Poon L, Syngelaki A, Mosimann B, Nicolaides KH. Fetal fraction in maternal plasma cell-free DNA at 11–13 weeks’ gestation: effect of maternal and fetal factors. Fetal Diagn Ther. 2012;31:237–43. doi: 10.1159/000337373. [DOI] [PubMed] [Google Scholar]
- 32.Ashoor G, Syngelaki A, Poon LC, Rezende JC, Nicolaides KH. Fetal fraction in maternal plasma cell-free DNA at 11–13 weeks’ gestation: relation to maternal and fetal characteristics. Ultrasound Obstet Gynecol. 2013;41:26–32. doi: 10.1002/uog.12331. [DOI] [PubMed] [Google Scholar]
- 33.Wang E, Batey A, Struble C, Musci T, Song K, Oliphant A. Gestational age and maternal weight effects on fetal cell-free DNA in maternal plasma. Prenat Diagn. 2013;33:662–6. doi: 10.1002/pd.4119. [DOI] [PubMed] [Google Scholar]
- 34.Lapaire O, Volgmann T, Grill S, Hosli I, Zanetti-Daellenbach R, Yan Zhong Xiao, et al. Significant correlation between maternal body mass index at delivery and in the second trimester, and second trimester circulating total cell-free DNA levels. Reprod Sci. 2009;16:274–9. doi: 10.1177/1933719108327599. [DOI] [PubMed] [Google Scholar]
- 35.Vora NL, Johnson KL, Basu S, Catalano PM, Hauguel-de Mouzon S, Bianchi DW. A multifactorial relationship exists between total circulating cell-free DNA levels and maternal BMI. Prenat Diagn. 2012;32:912–4. doi: 10.1002/pd.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Canick JA, Kloza EM, Lambert-Messerlian GM, Haddow JE, Ehrich M, van den Boom D, et al. DNA sequencing of maternal plasma to identify Down syndrome and other trisomies in multiple gestations. Prenat Diagn. 2012;32:730–4. doi: 10.1002/pd.3892. [DOI] [PubMed] [Google Scholar]
- 37.Srinivasan A, Bianchi DW, Liao W, Sehnert AJ, Rava RP. Maternal plasma DNA sequencing: effects of multiple gestation on aneuploidy detection and the relative cell-free fetal DNA (cffDNA) per fetus. Am J Obstet Gynecol. 2013;208:S31. [Google Scholar]
- 38.Jiang P, Chan KC, Liao GJ, Zheng YW, Leung TY, Chiu RW, et al. Fetal Quant: deducing fractional fetal DNA concentration from massively parallel sequencing of DNA in maternal plasma. Bioinformatics. 2012;28:2883–90. doi: 10.1093/bioinformatics/bts549. [DOI] [PubMed] [Google Scholar]
- 39.Leung TY, Qu JZ, Liao GJ, Jiang P, Cheng YK, Chan KC, et al. Noninvasive twin zygosity assessment and aneuploidy detection by maternal plasma DNA sequencing. Prenat Diagn. 2013;33:75–81. doi: 10.1002/pd.4132. [DOI] [PubMed] [Google Scholar]
- 40.Qu JZ, Leung TY, Jiang P, Liao GJ, Cheng YK, Sun H, et al. Noninvasive prenatal determination of twin zygosity by maternal plasma DNA analysis. Clin Chem. 2013;59:427–35. doi: 10.1373/clinchem.2012.194068. [DOI] [PubMed] [Google Scholar]
- 41.Chen EZ, Chiu RW, Sun H, Akolekar R, Chan KC, Leung TY, et al. Noninvasive prenatal diagnosis of fetal trisomy 18 and trisomy 13 by maternal plasma DNA sequencing. PLoS One. 2011;6:e21791. doi: 10.1371/journal.pone.0021791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mennuti MT, Cherry AM, Morrissette JJ, Dugoff L. Is it time to sound an alarm about false-positive cell-free DNA testing for fetal aneuploidy? [Epub ahead of print] Am J Obstet Gynecol. 2013 Mar 22; doi: 10.1016/j.ajog.2013.03.027. pii:S0002-9378(13)00301-3. [DOI] [PubMed] [Google Scholar]
- 43.Futch T, Spinosa J, Bhatt S, de Feo E, Rava RP, Sehnert AJ. Initial clinical laboratory experience in noninvasive prenatal testing for fetal aneuploidy from maternal plasma DNA samples. Prenat Diagn. 2013;33:569–74. doi: 10.1002/pd.4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fernando MR, Chen K, Norton S, Krzyzanowski G, Bourne D, Hunsley B, et al. A new methodology to preserve the original proportion and integrity of cell-free fetal DNA in maternal plasma during sample processing and storage. Prenat Diagn. 2010;30:418–24. doi: 10.1002/pd.2484. [DOI] [PubMed] [Google Scholar]
- 45.Benn P, Borell A, Chiu RW, Cuckle H, Dugoff L, Faas B, et al. Position statement from the Aneuploidy Screening Committee on behalf of the International Society for Prenatal Diagnosis. Prenat Diagn. 2013;33:622–9. doi: 10.1002/pd.4139. [DOI] [PubMed] [Google Scholar]
- 46.The American College of Obstetricians and Gynecologists Committee of Genetics and the Society for Maternal Fetal Medicine Publications Committee. Committee Opinion Number 545. Dec, 2012. [Google Scholar]
- 47.Langlois S, Brock J, Wilson RD, Audibert F, Brock J-A, Cartier L, et al. Current status in non-invasive prenatal detection of Down syndrome, trisomy 18, and trisomy 13 using cell-free DNA in maternal plasma. J Obstet Gynaecol Can. 2013;35:177–81. doi: 10.1016/S1701-2163(15)31025-2. [DOI] [PubMed] [Google Scholar]
- 48.Fairbrother G, Johnson S, Musci TJ, Song K. Clinical experience of noninvasive prenatal testing with cell-free DNA for fetal trisomies 21, 18, and 13, in a general screening population. Prenat Diagn. 2013;33:580–3. doi: 10.1002/pd.4092. [DOI] [PubMed] [Google Scholar]
- 49.Song Y, Liu C, Qi H, Zhang Y, Bian X, Liu J. Noninvasive prenatal testing of fetal aneuploidies by massively parallel sequencing in a prospective Chinese population. Prenat Diagn. 2013;33:700–6. doi: 10.1002/pd.4160. [DOI] [PubMed] [Google Scholar]
- 50.Gil MM, Quezada MS, Bregant B, Ferraro M, Nicolaides KH. Implementation of maternal blood cell-free DNA testing in early screening for aneuploidies. Ultrasound Obstet Gynecol. 2013;42:34–40. doi: 10.1002/uog.12504. [DOI] [PubMed] [Google Scholar]
- 51.Nicolaides KH, Wright D, Poon LC, Syngelaki A, Gil M. First-trimester contingent screening for trisomy 21 by biomarkers and maternal blood cell-free DNA testing. Ultrasound Obstet Gynecol. 2013;42:41–50. doi: 10.1002/uog.12511. [DOI] [PubMed] [Google Scholar]
- 52.Hahnemann JM, Vejerslev LO. Accuracy of cytogenetic findings on chorionic villus sampling (CVS)–diagnostic consequences of CVS mosaicism and non-mosaic discrepancy in centres contributing to EUCROMIC 1986–1992. Prenat Diagn. 1997;17:801–20. doi: 10.1002/(sici)1097-0223(199709)17:9<801::aid-pd153>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 53.Hall AL, Drendel HM, Verbrugge JL, Reese AM, Schumacher KL, Griffith CB, et al. Positive cellfree fetal DNA testing for trisomy 13 reveals confined placental mosaicism. Genet Med. 2013;15:729–32. doi: 10.1038/gim.2013.26. [DOI] [PubMed] [Google Scholar]
- 54.Pan M, Li FT, Li Y, Jiang FM, Li DZ, Lau TK, et al. Discordant results between fetal karyotyping and non-invasive prenatal testing by maternal plasma sequencing in a case of uniparental disomy 21 due to trisomic rescue. Prenat Diagn. 2013;33:598–601. doi: 10.1002/pd.4069. [DOI] [PubMed] [Google Scholar]
- 55.Lau TK, Jiang FM, Stevenson RJ, Lo TK, Chan LW, Chan MK, et al. Secondary findings from noninvasive prenatal testing for common fetal aneuploidies by whole genome sequencing as a clinical service. Prenat Diagn. 2013;33:602–8. doi: 10.1002/pd.4076. [DOI] [PubMed] [Google Scholar]
- 56.Choi H, Lau TK, Jiang FM, Chan MK, Zhang HY, Lo PS, et al. Fetal aneuploidy screening by maternal plasma DNA sequencing: “False positive” due to confined placental mosaicism. Prenat Diagn. 2013;33:198–200. doi: 10.1002/pd.4024. [DOI] [PubMed] [Google Scholar]
- 57.Yao HL, Zhang L, Zhang H, Jiang F, Hu H, Chen F, et al. Noninvasive prenatal genetic testing for fetal aneuploidy detects maternal trisomy X. Prenat Diagn. 2012;32:1114–6. doi: 10.1002/pd.3946. [DOI] [PubMed] [Google Scholar]
- 58.Osborne CM, Hardisty E, Devers P, Kaiser-Rogers K, Hayden MA, Goodnight W, et al. Discordant noninvasive prenatal testing results in a patient subsequently diagnosed with metastatic disease. Prenat Diagn. 2013;33:609–11. doi: 10.1002/pd.4100. [DOI] [PubMed] [Google Scholar]
- 59.Morris JK, Alberman E, Scott C, Jacobs P. Is the prevalence of Klinefelter syndrome increasing? Eur J Hum Genet. 2008;16:163–70. doi: 10.1038/sj.ejhg.5201956. [DOI] [PubMed] [Google Scholar]
- 60.Bianchi DW, Prosen T, Platt LD, Goldberg JD, Abuhamad AZ, Rava RP, et al. Massively parallel sequencing of maternal plasma DNA in 113 cases of fetal nuchal cystic hygroma. Obstet Gynecol. 2013;121:1057–62. doi: 10.1097/AOG.0b013e31828ba3d8. [DOI] [PubMed] [Google Scholar]
- 61.Mazloom AR, Dzakula Z, Oeth P, Wang H, Jensen T, Tynan J, et al. Noninvasive prenatal detection of sex chromosomal aneuploidies by sequencing circulating cell-free DNA from maternal plasma. Prenat Diagn. 2013;33:591–7. doi: 10.1002/pd.4127. [DOI] [PubMed] [Google Scholar]
- 62.Bischoff FZ, Lewis DE, Simpson JL. Cell-free fetal DNA in maternal blood: kinetics, source and structure. Hum Reprod Update. 2005;11:59–67. doi: 10.1093/humupd/dmh053. [DOI] [PubMed] [Google Scholar]
- 63.Samango-Sprouse C, Banjevic M, Ryan A, Sigurjonsson S, Zimmermann B, Hill M, et al. SNPbased non-invasive prenatal testing detects sex chromosome aneuploidies with high accuracy. Prenat Diagn. 2013;33:643–9. doi: 10.1002/pd.4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guex N, Iseli C, Syngelaki A, Deluen C, Pescia G, Nicolaides KH, et al. A robust second-generation genome-wide test for fetal aneuploidy based on shotgun sequencing cell-free DNA in maternal blood. Prenat Diagn. 2013;33:707–10. doi: 10.1002/pd.4130. [DOI] [PubMed] [Google Scholar]
- 65.Wapner RJ, Martin CL, Ballif BC, Eng CM, Zachary JM, Savage M, et al. Chromosomal microarray versus karyotyping for prenatal diagnosis. N Engl J Med. 2012;367:2175–84. doi: 10.1056/NEJMoa1203382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Peters D, Chu T, Yatsenko SA, Hendrix N, Hogge WA, Surti U, et al. Noninvasive prenatal diagnosis of a fetal microdeletion syndrome. N Engl J Med. 2011;365:1847–8. doi: 10.1056/NEJMc1106975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jensen TJ, Dzakula Z, Deciu C, van den Boom D, Ehrich M. Detection of microdeletion 22q11. 2 in a fetus by next-generation sequencing of maternal plasma. Clin Chem. 2012;58:1148–51. doi: 10.1373/clinchem.2011.180794. [DOI] [PubMed] [Google Scholar]
- 68.Chen S, Lau TK, Zhang C, Xu C, Xu Z, Hu P, et al. A method for noninvasive detection of fetal large deletions/duplications by low coverage massively parallel sequencing. Prenat Diagn. 2013;33:584–90. doi: 10.1002/pd.4110. [DOI] [PubMed] [Google Scholar]
- 69.Yu S, Jiang P, Choy KW, Chan KC, Won H-S, Leung WC, et al. Noninvasive prenatal molecular karyotyping from maternal plasma. PLoS One. 2013 Apr 17; doi: 10.1371/journal.pone.0060968. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Srinivasan A, Bianchi DW, Huang H, Sehnert AJ, Rava RP. Noninvasive detection of fetal sub chromosome abnormalities via deep sequencing of maternal plasma. Am J Hum Genet. 2013;92:167–76. doi: 10.1016/j.ajhg.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Snyder MW, Simmons LE, Kitzman JO, Santillan DA, Santillan MK, Gammill HS, et al. Noninvasive fetal genome sequencing: a primer. Prenat Diagn. 2013;33:547–54. doi: 10.1002/pd.4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kitzman JO, Snyder MW, Ventura M, Lewis AP, Qiu R, Simmons LE, et al. Noninvasive whole-genome sequencing of a human fetus. Sci Transl Med. 2012;4:137ra76. doi: 10.1126/scitranslmed.3004323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fan HC, Gu W, Wang J, Blumenfeld YJ, El-Sayed YY, Quake SR. Non-invasive prenatal measurement of the fetal genome. Nature. 2012;487:320–4. doi: 10.1038/nature11251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chetty S, Garabedian MJ, Norton ME. Uptake of noninvasive prenatal testing (NIPT) in women following positive aneuploidy screening. Prenat Diagn. 2013;33:542–6. doi: 10.1002/pd.4125. [DOI] [PubMed] [Google Scholar]
- 75.Benn P, Cuckle H, Pergament E. Non-invasive prenatal diagnosis for Down syndrome: the paradigm will shift, but slowly. Ultrasound Obstet Gynecol. 2012;39:127–30. doi: 10.1002/uog.11083. [DOI] [PubMed] [Google Scholar]