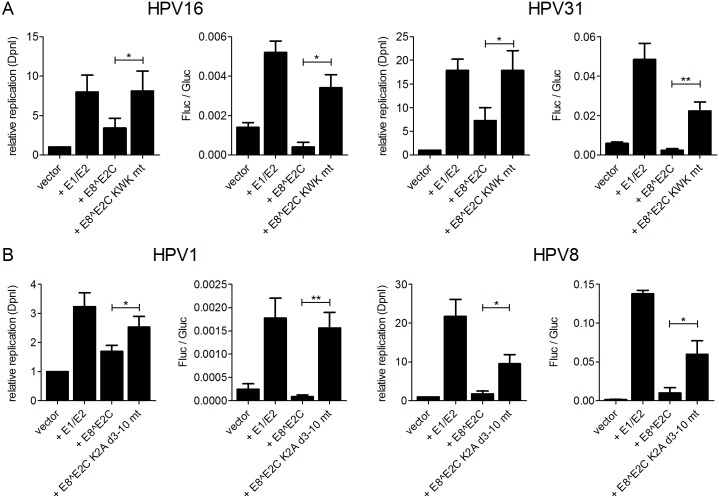
Fig 8. E8^E2C proteins inhibit replication in a reporter-based replication assay.
(A) Replication assay for HPV16 and HPV 31. HeLa cells were transiently transfected with a reporter plasmid containing the HPV16 or 31 URR and the viral early promoter driving the firefly luciferase gene (10 ng) (pGL 16 URR luc or pGL 31 URR luc) and expression vectors for E1 (100 ng) (pSG16 E1 or pSG31 E1), E2 (10 ng) (pSG16 E2 or pSX31 E2) and E8^E2C or the E8^E2C KWK mt (HPV16: 1 ng; HPV31: 10 ng) (pSG31 E8^E2C, pSG31 E8 KWK mt, pSG16 E8^E2C, pSG16 E8 KWK mt). (B) Replication assays for HPV1 and HPV8. RTS3b cells were transfected with the HPV1 reporter plasmid and expression vectors amounts described for HPV31 (pGL 1 URR luc, pSG 1 E1, pSG 1 E2, pSG 1 E8^E2C, pSG1 E8^E2C K2A d3-10 mt). RTS3b cells were transfected with the HPV8 reporter plasmid and expression vectors as described for HPV16 (pGL 8 URR luc, pSG 8 E1, pSG 8 E2, pSG 8 E8^E2C, pSG 8 E8^E2C K2A d3-10 mt). Differences in the amount of DNA were adjusted with the empty vector (pSG5). pCMV-gluc (0.3ng) was used as an internal control. Replication activity is shown as the relative replication measured by real-time PCR after DpnI-digestion (“relative replication (DpnI)”) (reporter plasmid cotransfected with the empty expression vector = “vector” is set to 1) or as the ratio between firefly luciferase (fluc) to gaussia luciferase (gluc) activities (“Fluc/Gluc”). Data are derived from at least three independent experiments performed in duplicate. Error bars indicate the standard errors of the means. A paired two tailed t-test was used to determine statistical significance (*p<0.05, **p<0.01).