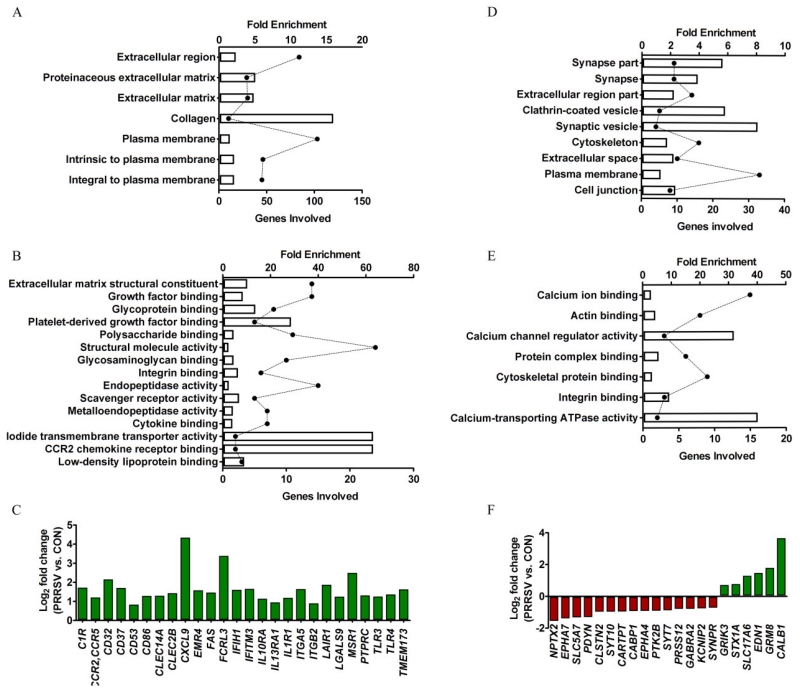
Figure 2. Gene ontology (GO) functional enrichment analysis using DAVID bioinformatics 6.7 (A, B, D, E) and clusters of sensome (C) and synaptic genes (F).
(A, B) Top enriched GO biological process (A) and molecular function (B) associated with up-regulated DEGs (bars correspond to fold enrichment; solid circles correspond to number of gene involved; statistics for enrichment, Bonferroni-adjusted P < 0.05). (C) Manually identified DEGs that are part of the microglia sensome. (D, E) Top enriched GO cellular component (D) and molecular function (E) associated with down-regulated DEGs (statistics for enrichment, non-adjusted P < 0.05). (F) Manually identified DEGs encoding synaptic proteins.