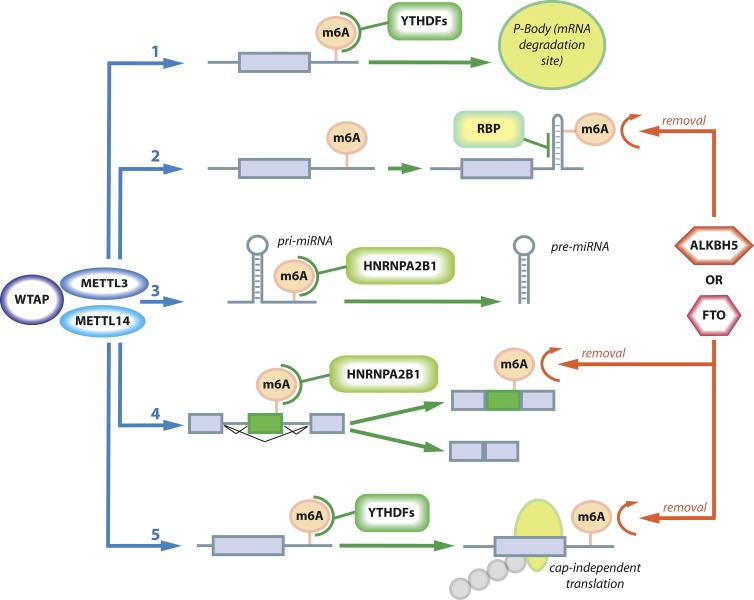
Figure 2.
The dynamic interplay of factors writing, reading, and erasing m6A modifications. The m6A mark is written by METTL3 or METTL14 (blue) in complex with WTAP (purple), whereas the proteins ALKBH5 or FTO (red) are erasers of m6A modifications. Several reading mechanisms have been characterized: (1) YTHDF proteins (green) can detect the m6A mark and shift localization of the transcripts to P-bodies and thereby promote degradation; (2) m6A can cause structural changes and thereby lead to differential binding of RNA-binding proteins (RBP, green); (3 and 4) HNRNPA2B1 (green) can bind to m6A sites in pri-miRNAs or pre-mRNAs and either promote maturation of miRNAs (3) or cause alternative splicing (4); and finally, (5) under stress conditions, m6A can also cause cap-independent translation. m6A writing mechanisms are in blue, reading mechanisms are in green, and erasing mechanisms are in red.