Fig. 5.
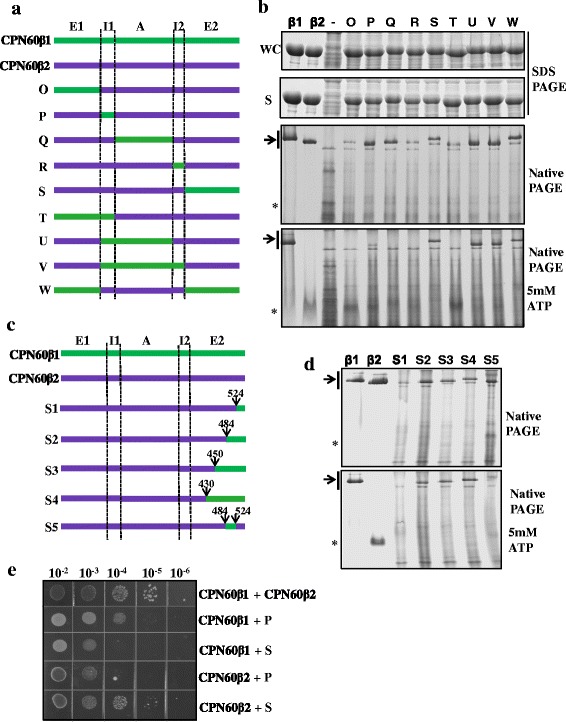
Segments influencing CPN60β homo-oligomer disassembly. a Diagram of chaperonin chimeras between CPN60β1 and CPN60β2. The domain designation and amino acid numbering are the same as described for Fig. 3a. b Oligomer formation and disassembly of chaperonin chimeras upon incubation with ATP. Visualization of the disassembly of CPN60β chimeric oligomers was performed as described in Fig. 3c. c Diagram of CPN60β subunits and the constructed chaperonin chimeras. The swapping positions are indicated in the figure. d Disassembly of chaperonin chimeras. The untreated soluble fractions and soluble fractions treated with 5 mM ATP/Mg were resolved by 6 % native PAGE and visualized by Coomassie staining. Recombinantly induced CPN60β1 and CPN60β2 oligomers were loaded as controls. The positions of oligomers and monomers are indicated by an arrow and *, respectively. e Functional replacement of GroEL by coexpression of CPN60β with chimeras. CPN60β chimeras and GroES were expressed in GroEL/ES-deficient E. coli strain MGM100. The strains were grown on medium supplemented with glucose and IPTG at 37 °C for 24 hours
