Fig. 1.
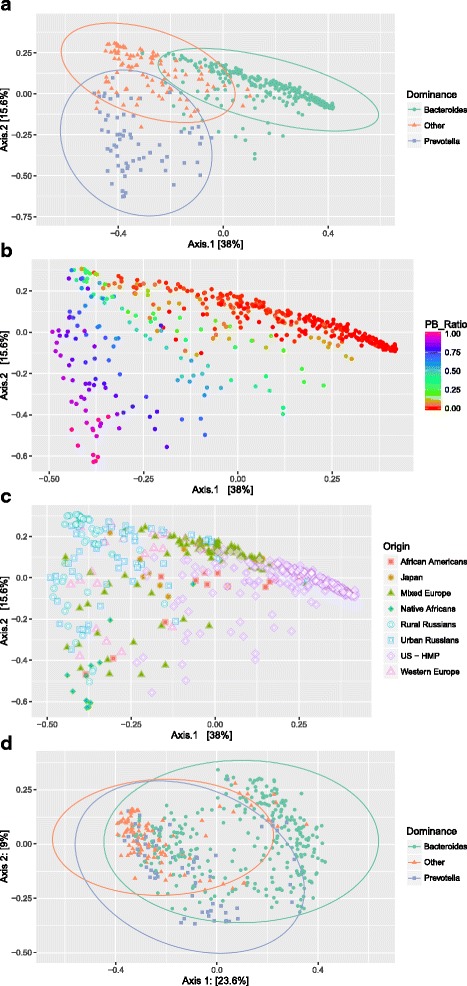
PCoA plots using the Bray distance metric with all the samples except for the Yatsunenko study. a Samples colored by their most prominent taxon. If the sample is dominated neither by Prevotella nor Bacteroides, it is classified as other. Ellipses were projected for each group in the plot. The ellipse axes represent the directions of the within-group covariance matrices, and their bounds represent two standard deviations in each direction from the cluster mean. b Samples are colored by their value for the Prevotella ratio (relative abundance of Prevotella/[Bacteroides + Prevotella]) on a spectrum with red indicating no Prevotella and purple no Bacteroides. c Samples are colored by population of origin. d The Bray distance has been recalculated without the relative abundances of Bacteroides and Prevotella. Samples are colored by most prominent taxon in the original samples distributions, and ellipses were projected for each group in the plot (same as in plot a)
