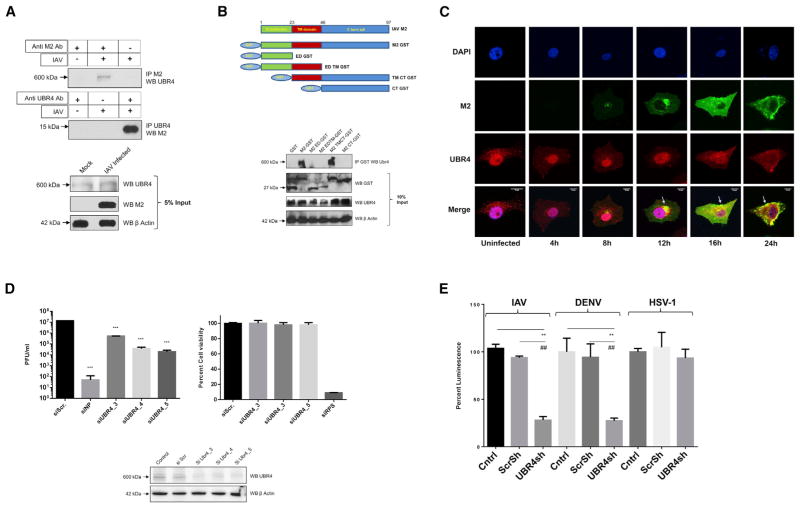
Figure 4. UBR4 Interacts with the M2 Ion Channel and Is Required for IAV Replication.
(A) A549 cells were infected with A/WSN/33 (MOI = 2) for 24 hr and lysed in IP buffer. Lysates were subjected to immunoprecipitation using antibodies against M2 and UBR4. Immunoprecipitated protein samples and 5% input were subjected to SDS-PAGE/western blotting using indicated antibodies.
(B) N-terminal GST-tagged deletion constructs of A/WSN/33 M2 (top) were transfected in HEK293T cells. At 48 hr later, cells were lysed and subjected to immunoprecipitation with anti-GST antibody. Immunoprecipitated samples and 10% input were subjected to SDS-PAGE/western blotting (bottom).
(C) A549 cells were infected with A/WSN/33 (MOI = 2), and cells were fixed at indicated time points for immunostaining. Nuclei are depicted in blue, M2 in green, and UBR4 in red. Arrows allocate M2-UBR4 co-localization. Images of three representative independent experiments are shown. Scale bar represents 10 μm.
(D) A549 cells were transfected with siNP, siUBR4, or siScr; 48 hr later, cell viability and UBR4 expression levels were determined. Alternatively, cells were infected with A/WSN/33 (MOI = 0.01) for 24 hr, and supernatants were titered. Shown is one of two independent duplicate experiments ± SD.
(E) UBR4 stable knockdown or control A549 cells were infected with IAV luciferase (MOI = 0.2), Dengue Luciferase, or Herpes 1 luciferase reporter virus (MOI = 0.1); 48 hr later cells were lysed, and luciferase activity was measured. The mean luminescence values ± SD of three independent experiments relative to control were plotted. * and # indicate p values compared with control and Scr shRNA, respectively.
See also Figures S4 and S5.