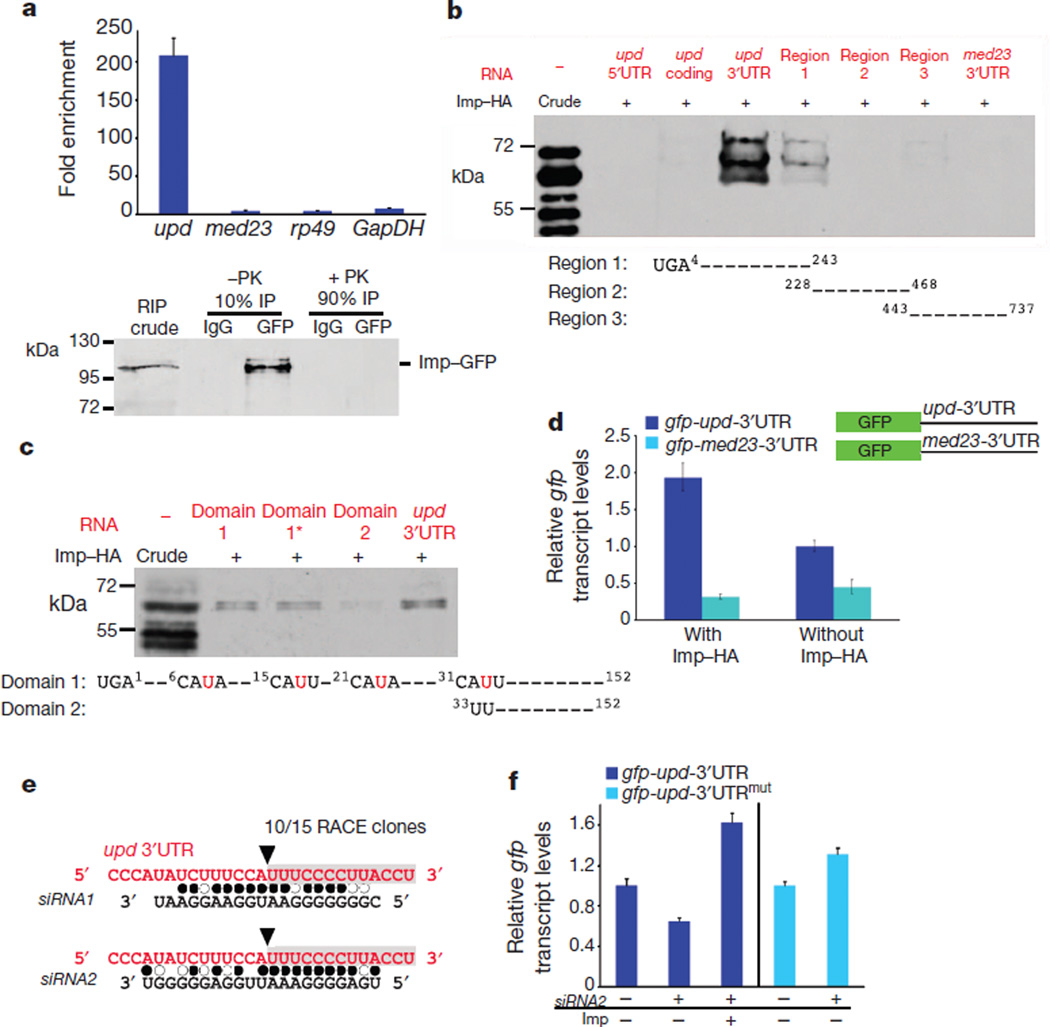
Figure 2. Imp binds to upd mRNA and counteracts siRNA-mediated degradation.
a, Top, fold-enrichment of the indicated mRNAs after RNA immunoprecipitation (RIP) with anti-GFP antibodies relative to control IgG from the testes of young Imp–GFP flies. Bottom, immunoblot for Imp–GFP. Ten per cent of the immunoprecipitate was used to confirm immunoprecipitation efficiency; the remaining 90% was treated with proteinase K (PK) and used for qRT–PCR. b, c, In vitro protein–RNA binding assay. Top panels, immunoblots for Imp–HA binding to designated RNA fragments. Imp–HA binds to region 1 of the upd 3′UTR (b), and to domain 1 and 1* within region 1 of the upd 3′UTR, but not to domain 2 (c). Mutations in domain 1* of the upd 3′UTR are indicated in red (c). d, qRT–PCR showing relative gfp mRNA levels in cells co-transfected with actin-GAL4 and the indicated gfp reporter constructs. Expression of gfp is presented relative to cells without Imp–HA and is normalized toGAL4. Note that gfp-upd-3′UTR mRNA is sixfold higher than gfp-med23-3′UTR in cells expressing Imp–HA. e, Sequence of cleavage product from aged testes identified a fragment starting at nucleotide 33 within the upd 3′UTR (black arrowhead, grey box). Alignment of the upd 3′UTR with endo-siRNAs found in aged testis library. Filled circles, canonical base pairing; open circles, GU pairs. f, qRT–PCR showing gfp fromS2 cells transfected with gfp-upd-3′UTR or with gfp-upd-3′UTRmut (32AUU = CGG), with or without Imp and siRNA2. Levels normalized to rp49 and relative to control gfp levels without siRNA2. Error bars in a, d and f denote s.d. of triplicate measurements.