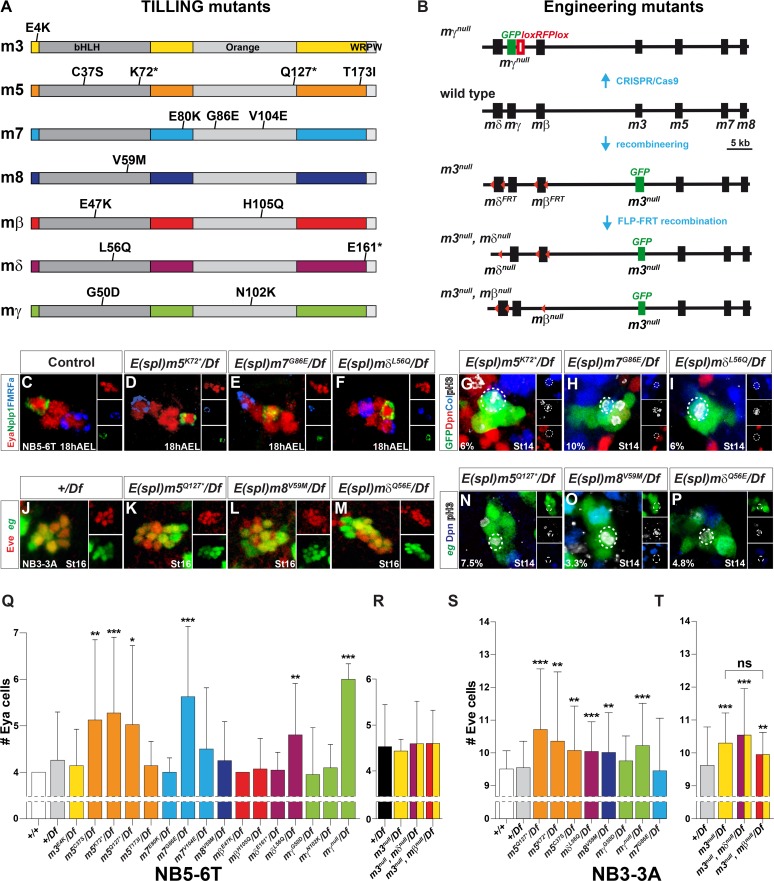
Fig 3. Single and differential E(spl)-HLH gene function in neuroblasts.
(A-B) Outline of E(spl)-HLH TILLING, CRISPR/Cas9 and recombineering mutants. (C) Control, showing the four Ap cells (Eya+) in T2-T3. (D-F) Extra Ap cells, and (G-I) aberrantly dividing NB5-6T daughter cells; in m5K72*, m7G86E and mδQ56E, when placed over deletion Df(3R)BSC751 (G-I; percentages denote NB5-6T lineages with a dividing daughter cell). (J-M) Extra Eve cells, and (N-P) aberrantly dividing NB3-3A daughter cells; in m5Q127*, m8V59M and mδQ56E, when placed over deletion Df(3R)BSC751 (percentages denote NB3-3A lineages with a dividing daughter cell). (Q-T) Quantification of phenotypes. Four E(spl)-HLH genes show significant effects in NB5-6T, while five show effects in NB3-3A. Notably, m7 only affects NB5-6T, while m3 and m8 only affects NB3-3A (* p≤0.05, ** p≤0.01, *** p≤0.001; +/-SD; Kruskal-Wallis, Dunn’s posthoc; n≥22 lineages). TILLING alleles and the CRISPR/Cas9 mγnull allele were placed over deletion Df(3R)BSC751. The recombineering alleles; m3null, mδnull and mβnull, were deleted in a BAC, inserted on chromosome 2 (51D) and crossed into a Df(3R)gro32.2, P-gro/(E(spl)-CΔmδ-m6 genetic background. Control was Df(3R)gro32.2, P-gro/+.