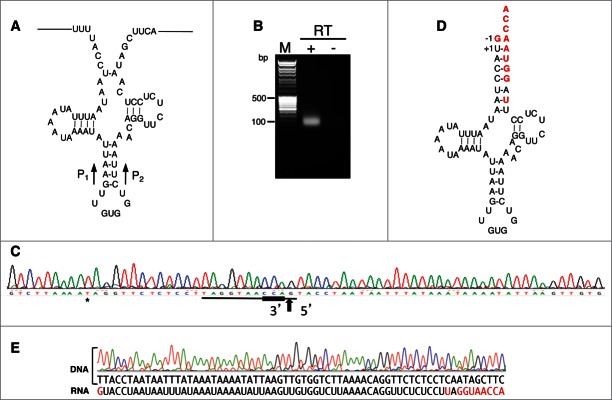
Figure 2.
The biogenesis of A. vulgare mt tRNAHis requires repair at both the 5′- and 3′-ends. (A) Inferred tRNA secondary structure of the tRNAHis primary transcript deduced from the genomic sequence. The primers P1/P2 used for cRT-PCR analyses are depicted by black arrows, their precise positions are presented on Table S2. (B) Total nucleic acids were analyzed by cRT-PCR using primers P1 and P2. After circularization in the presence of T4 RNA ligase, cDNA was synthezized in the presence of primer P1. The image shows the ethidium bromide stained gel of the PCR product amplified using primers P1 and P2. The lane marked M shows the migration of the DNA ladder. (C) Sample sequence of one of the clones obtained from the cRT-PCR product shown in (B). The 3′- and 5′- extremities of the tRNA molecule are presented. The two extremities are separated by a vertical arrow. The CCA extremity is underlined in bold. Note that primers used for tRNA-His sequencing (Table S2) were designed from the online reference genome sequence (GenBank accession EF643519.3), which presents one nucleotide polymorphism in the variable loop (indicated by an asterisk). This did not affect the amplification of cDNA and the demonstration of post-transcriptional modification. (D) Inferred secondary structure of the mature tRNAHis deduced from the sequence of the cloned cRT-PCR product. Nucleotides differing between the genomic sequence and the RNA sequence are indicated in red. (E) Comparison between the sequence of the genomic DNA region coding for the A.vulgare mt tRNAHis and the sequence deduced from the corresponding RNA amplified by cRT-PCR.