Figure 1.
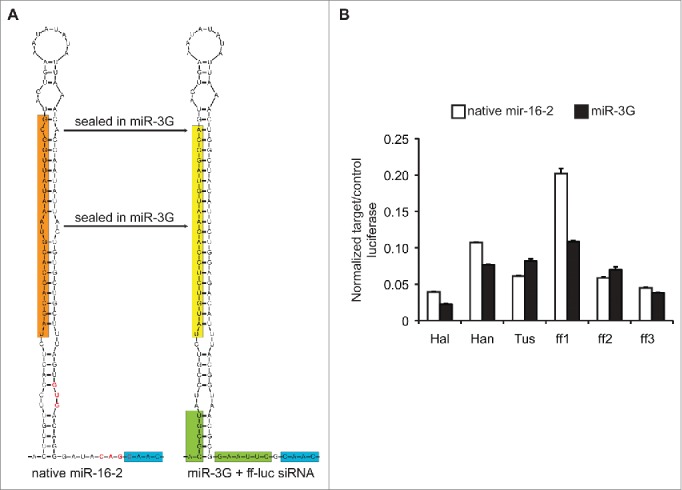
Structure and function of miR-16-2-based RNAi triggers. (A) Comparison of the native miR-16-2 hairpin context relative to the miR-3G format. The mature, primary, 5p miR-16-2 sequence is highlighted in orange, whereas the targeting (antisense) sequence derived from miR-3G is highlighted in yellow. The novel MluI (ACGCGU) and EcoRI (GAAUUC) sites for cloning unique hairpins into miR-3G are highlighted in green. Note, the native miR-16-2 context contains a GHG motif and 2 putative CNNC miRNA processing enhancers, red text and blue highlight. The sequences in red text, including the GHG motif and one of the putative CNNC motifs, has been modified in miR-3G so as to incorporate the EcoRI cloning site. Structures were predicted using the default settings from the mFold web server (http://unafold.rna.albany.edu/?q = mfold/rna-folding-form).50 These are partial sequence views, and structures of the full ∼175 nt contexts are available in Supplemental Figure 2. (B) Knockdown efficiency of distinct targeting sequences within the native miR-16-2 (black bars) or miR-3G (white bars) hairpin contexts. ff-luc (target) was normalized to Rr-luc (control) luciferase expression, and all amiRNAs were co-transfected in a 1:30 w/w ratio to the target plasmid. Knockdown was measured ∼40 hour post transfection into HEK293T cells. Shown is a representative experiment (N = 2 ) performed in technical triplicate. Error bars represent one standard deviation of uncertainty, based on the sampled triplicates, and bars represent the mean for individual data sets.
