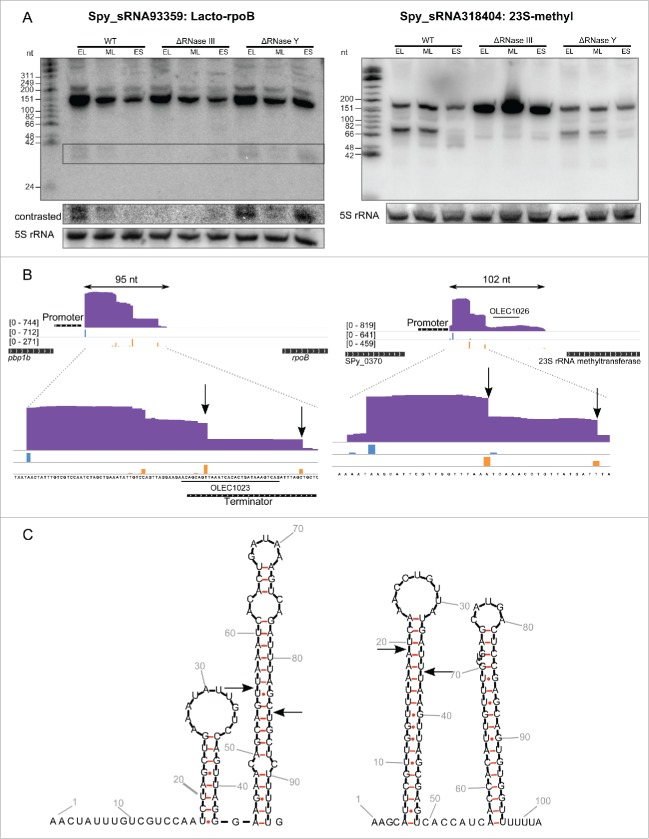
Figure 1.
For figure legend see page .Figure 1. (see previous page) Lacto-rpoB and 23S methyl RNA elements are regulated by RNase III in S. pyogenes. A. Northern blot analysis (polyacrylamide gel electrophoresis) of Lacto-rpoB and 23S-methyl RNA expression in WT (SF370), ΔRNase III (Δrnc) and ΔRNase Y (Δrny) strains grown to early logarithmic (EL), mid logarithmic (ML) and early stationary (ES) phases. 5S rRNA is used as loading control. B. Expression profiles of Lacto-rpoB and the 23S-methyl motif with surrounding genes captured using the Integrative Genomics Viewer (IGV) software. The sequence coverage was calculated using BEDTools-Version-2.15.0 and the scale is given in number of reads per million. The distribution of reads starting (5') and ending (3') at each nucleotide position is represented in blue and orange, respectively. The position of the oligonucleotide probes (OLEC) used in Northern blot analysis is indicated. C. Prediction of RNA secondary structure using RNAfold (rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi). The arrows represent putative cleavages by RNase III (nucleotides determined by analyzing the 5' and 3' ends of the sRNAs in sRNA sequencing data).