Figure 1.
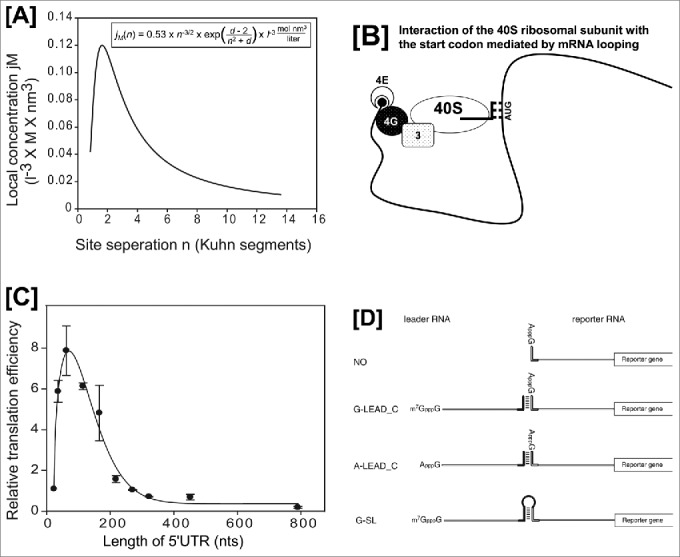
Evidence for cap-dependent translation mediated by RNA-looping. Panels [A] and [C] are modified versions of Fig. 5B and 5A from reference 2. [A] Graph depicting the relationship between the local concentration, jM, and the distance, n (number of Kuhn segments), predicted by the equation in the box. Because the experiments required to obtain the parameter d cannot be performed, we used d = 0 in the calculation.1 The peak of the graph occurred at n = 1.62. [B] Schematic diagram of the interaction between the AUG start codon and a 40S ribosome associated with the 5′-cap structure. It should be noted that the intervening RNA segment is looped out from the complex composed of the 43S ribosome and the start codon. Abbreviations: 4E, eIF4E; 4G, eIF4G; 3, eIF3; and 40S, 40S ribosomal subunit. The initiator tRNAi associated with the 40S ribosome is depicted as lines. [C] Relative translation efficiencies of mRNAs harboring 5′UTRs of various lengths. Reporter DNAs (kindly provided by Dr. Vincent Mauro, The Scripps Research Institute, La Jolla, CA) were transfected into HEK293T cells, and luciferase activity was measured 24 h after transfection. [D] A modified version of Fig. 1 from reference 3. Schematic diagram depicting the leader and reporter RNAs. The reporter RNA contains a reporter gene (firefly luciferase). The leader RNAs are either m7GpppG-capped (G-LEAD_C) or ApppG-capped (A-LEAD_C). The leader RNAs (G-LEAD_C and A-LEAD_C) contain a 3′ nucleotide sequence that is complementary (solid bar) to the 5′ sequence in the reporter RNAs. An RNA complex containing both leader and reporter RNAs covalently connected by a stem-loop structure (i.e., within a single molecule) and with an m7GpppG-cap (G-SL) was constructed to serve as a control mRNA.
