Figure 2.
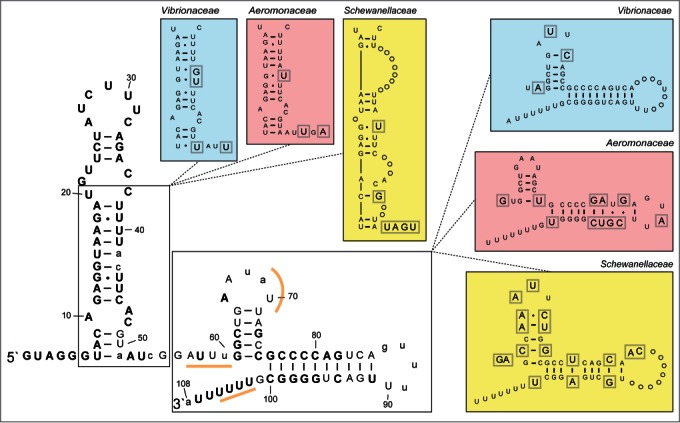
Secondary structure consensus model of the Spot 42 RNA. The structure model was made by aligning all known spf sequences, and by mapping the consensus sequence onto a secondary structure model of the E. coli Spot 42 (based on Møller et al.1). The structure consists of a relatively long 5` hairpin, a 9 nt long single-stranded region followed by a second hairpin and a rho-independent terminator. Level of identity is shown using different type of letters in the structure. Uppercase bold letters indicate 80–100 % identity, uppercase regular letters indicate 60–79% identity, and lowercase letters indicate <60% identity. Structural segments with family-specific (i.e., Vibrionaceae, Aeromonadaceae and Shewanellaceae) variations are shown in separate colored boxes. Here, circles indicate U or A insertions (compared to the "consensus"). Gray square around a letter symbolizes aberration from the consensus structure.
