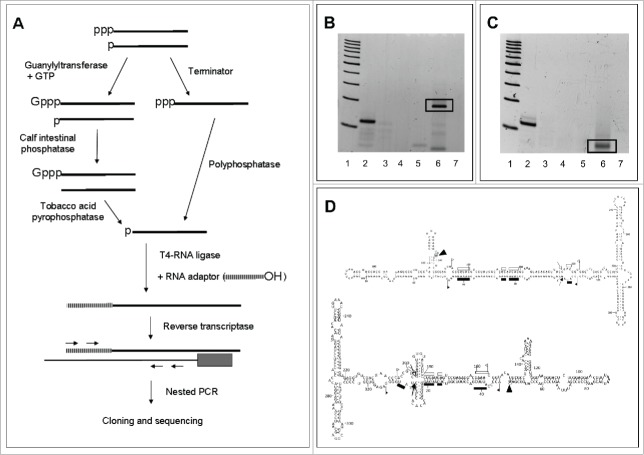
Figure 5.
Identification of the transcription initiation sites of ELVd strands. (A) Scheme of the 2 RLM-RACE approaches used to exclusively amplify RNAs with a 5′-triphosphorylated end. (B) Non-denaturing PAGE of the RT-PCR products resulting from applying in vitro capping and RLM-RACE to ELVd (+) strands. Lane 1, ladder of 100-bp DNA multimers. Lane 2, artificial reaction mixture containing an in vitro ELVd (+) transcript starting at position 210 combined with excesses of a CF11-fractionated RNA preparation from mock-inoculated eggplant. Lanes 3 and 4, reaction mixtures in which in vitro capping and addition of the RNA adaptor were omitted, respectively. Lanes 5 and 6, RNA preparations from mock-inoculated and ELVd-infected eggplant, respectively. Lane 7, reaction mixture in which the RNA template was omitted. Gel was stained with ethidium bromide. (C) Non-denaturing PAGE of the RT-PCR products resulting from applying RLM-RACE to ELVd (−) strands after their pretreatment with Terminator™ exonuclease and Polyphosphatase™. Lane 1, ladder of 100-bp DNA multimers. Lane 2, artificial reaction mixture containing an in vitro ELVd (−) transcript starting at position 113 combined with excesses of a CF11-fractionated RNA preparation from mock-inoculated eggplant. Lanes 3 and 4, reaction mixtures in which the treatment order with Terminator™ and Polyphosphatase™ was reversed, or the addition of the RNA adaptor was omitted, respectively. Lanes 5 and 6, RNA preparations from mock-inoculated and ELVd-infected eggplant, respectively. Lane 7, reaction mixture in which the RNA template was omitted. Gel was stained with ethidium bromide. (D) Arrowheads indicate the transcription initiation sites in the proposed in vivo conformations of ELVd (+) and (−) RNAs (upper and lower panels, respectively). Other details as in the legends to Figs. 1 and 2.