Figure 5.
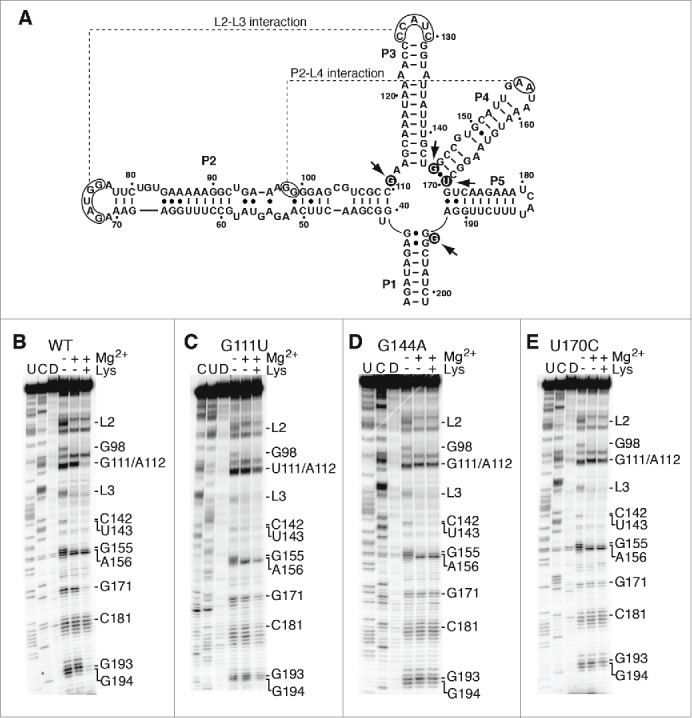
Junction core mutations are not important for the global folding of the aptamer. (A) Secondary structure representing the lysC aptamer. The regions participating in the formation of L2-L3 and P2-L4 interactions are indicated (dotted lines). Arrows indicate nucleotides involved in the corresponding tertiary interactions. Residues mutated are shown in black circles. The nucleotide nomenclature is based on previous studies.26,27 (B, C, D and E) SHAPE modification of selected lysC aptamers performed in the context of 10 mM NaCl. Experiments were done as a function of 10 mM Mg2+ and 5 mM lysine and analysis was performed using the wild-type aptamer (B) and variants G111U (C), G144A (D) and U170C (E). In each case, cytosine (C) and uracil (U) sequencing lanes were added to resolve the sequence. NMIA was replaced by DMSO (D) in control reactions. Nucleotides or regions exhibiting changes either in Mg2+ or lysine are indicated on the right.
