Figure 1.
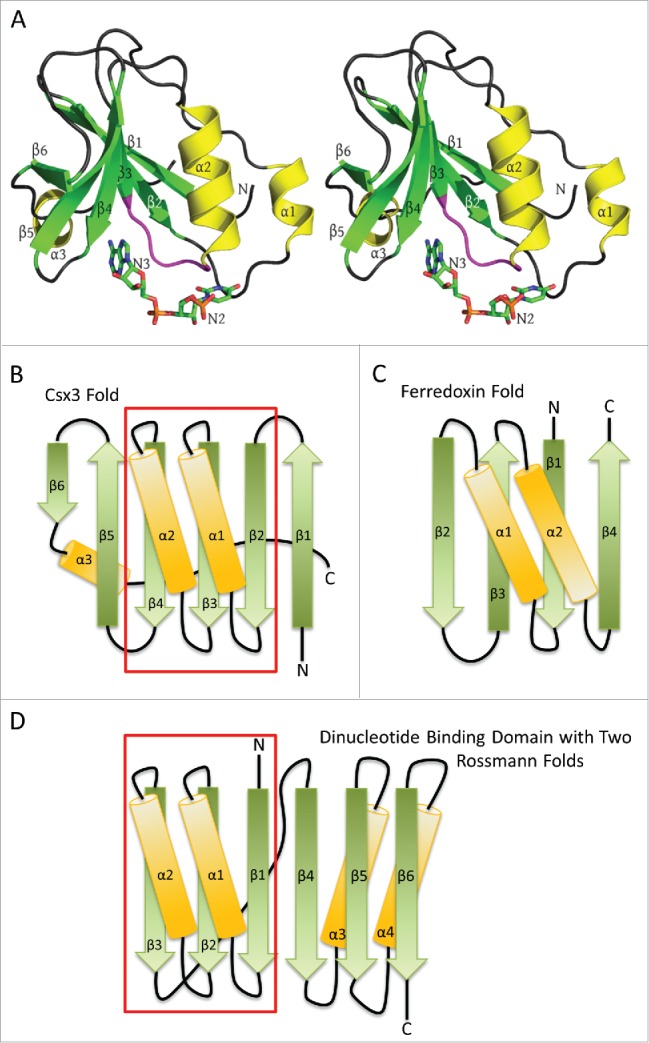
RNA recognition by AfCsx3. A) Stereo ribbon diagram of AfCsx3 Chain A bound to a ssRNA (PDB ID: 3WZI). Beta strands, α helices and the conserved β3-α2 loop (Ser44-Ile49) are shown in green, yellow and magenta, respectively. Secondary structures are labeled in order of their occurrence in the amino acid sequence. The orientation is roughly equivalent to that of Fig. 1 in Yan et al.,1 who modeled nucleotides N2 and N3 into subunit A which are shown as “sticks” and labeled accordingly. B) A topology or fold diagram of Csx3. Beta strands and α helices are colored according to panel A. Gradients are added to show the N-to-C directionality of the individual secondary structures, morphing from dark to lighter colors. The boxed region indicates the Rossmann fold present in Csx3. C) The similarly depicted RRM or ferredoxin-like fold. This fold is substantially different from the Csx3 fold in panels A and B, where Csx3 lacks the 4-stranded antiparallel β-sheet and the antiparallel helices of the RRM fold. D) Fold of the classic dinucleotide binding domain composed of 2 successive Rossmann folds. The first Rossmann fold is contained within the boxed region and is aligned with the Rossmann fold present in Csx3 in order to highlight the similarities between the 2 folds. Helix α3 of Csx3 runs along the back side of the mixed β sheet, contacting strands β4, β3 and β2, but has been slightly repositioned in panel B in order to accentuate the similarities between the Rossmann folds present in the classic dinucleotide binding domain and that in Csx3.
